Présentation

SHERLOCK4HAT
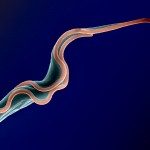
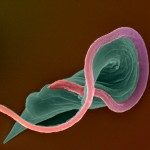
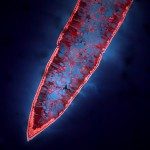
African trypanosomes are transmitted by the bite of the tsetse fly and cause the debilitating, and often fatal, neglected tropical disease sleeping sickness, or Human African Trypanosomiasis (HAT). Trypanosoma brucei gambiense, the parasites responsible for 98% of human cases, first reside in the patient blood and skin for months to years before invading the central nervous system, where they cause the neurological symptoms of the disease. HAT is approaching elimination, with the number of cases reported in 2017 dropping to approximately 1,442 from only a dozen African countries. In this context, HAT was included in the WHO roadmap on neglected tropical diseases, with 2020 set as target date for elimination as a public health problem. A secondary goal of zero transmission by 2030 has also been set. These targets have, in part, been encouraged by the success of surveillance efforts that rely on detecting extracellular trypanosomes in human blood. Nevertheless, the reduction in case numbers brings about other challenges. For example, the sensitivity of any diagnostic test diminishes as the disease burden drops, and this is being seen with the serological tests available for HAT. In this context, new highly sensitive and specific diagnostic tools will be required to accurately monitor the occurrence of new cases and the possible emergence of drug-resistant trypanosomes during the elimination phase. Most diagnostic tests currently under development are based on optimization of existing methods that may not combine all the requirements to stand up to the harsh constraints imposed by the elimination phase requirements, especially in terms of sensitivity and specificity.
We propose that adapting the recently developed Specific High-sensitivity Enzymatic Reporter unLOCKing (SHERLOCK) technology that combines a CRISPR-Cas system and lateral flow test to trypanosomes will provide the sensitivity and specificity required for a diagnostic test in the elimination and post elimination phases. The SHERLOCK system relies on the collateral effect of Cas13a promiscuous RNA cleavage activity upon target recognition. Combining the collateral effect with pre-amplification of RNAs resulted in rapid RNA detection with attomolar (10-18 moles/l) sensitivity and single-base mismatch specificity, in a diagnostic setting. This technology has been used to detect specific strains of Zika and Dengue viruses and distinguish pathogenic bacteria in a mixed sample. Furthermore, SHERLOCK reaction reagents can be lyophilized for cold-chain independence and long-term storage and be readily reconstituted on paper for field applications. A lateral flow test for a simple and rapid readout can be easily implemented after a reaction that does not exceed two hours from the sampling step.
The 5 partners of this IP-PTR 2019 project from IP Paris and IP Guinea propose to adapt, optimize and validate a new SHERLOCK approach for the detection of trypanosome-specific nucleic acid sequences in human body fluids. In total, SHERLOCK4HAT would provide an ideal field-adapted method for the diagnosis of HAT during the elimination and post-elimination phases.