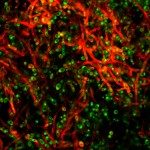
Présentation
Biofilms formed by C. albicans on indwelling devices play a major role in the persistence of nosocomial infections, largely because of their elevated tolerance to antifungals. The Fungal Biology and Pathogenicity Unit at Institut Pasteur has revealed a number of C. albicans genes that show increased expression upon biofilm formation. In particular, we have shown that a hypoxic environment forms within C. albicans biofilms, and that adaptation to hypoxia, (such as by increasing glycolysis) is necessary for efficient biofilm formation. More recently, we have developed and used a collection of approximately 600 C. albicans barcoded overexpression strains to implement a signature-tagged mutagenesis (STM) strategy to identify genes whose overexpression would confer a differential ability to participate in a multi-strain biofilm. Strikingly, most of the genes that we have identified encode cell wall glycosylphosphatidylinositol (GPI)-anchored proteins, the majority of which had no known role in biofilm formation previously. A similar approach has now been used to investigate colonization of the gastro-intestinal (GI) mucosa in animal models, as this represents an early step in the development of disseminated candidiasis. Our current research focuses on extending our STM approach to the entire C. albicans ORFeome. This is possible thanks to “The C. albicans ORFeome project” that we develop in collaboration with the Munro group at the University of Aberdeen and aims at establishing collections of overexpression plasmids and C. albicans strains for all C. albicans ORFs. We also intend to use our collection of genome-sequenced C. albicans isolates to identify genes with previously unknown roles in the colonization of abiotic and biotic surfaces. We are presently characterizing the cell wall proteins potentially involved in biofilm formation on abiotic surfaces, and the genes and proteins regulating gastro-intestinal colonization by C. albicans.