About
Hepatitis C virus E2 envelop glycoprotein
The hepatitis C virus (HCV) is a major cause of chronic liver disease worldwide, leading to cirrhosis and hepatocellular carcinoma. In spite of being the focus of intense research efforts, no vaccine is available against HCV, and current therapeutic treatments have limited efficacy and significant side effects. In particular, little is known about the structure of the envelope glycoproteins of this major human pathogen, in spite of their essential role in the viral cycle.
To obtain more structural information, we developed an efficient production system of recombinant E2 ectodomain (E2e), truncated immediately upstream its trans-membrane (TM) region, using Drosophila melanogaster cells. This system yields a majority of monomeric protein, which can be readily separated chromatographically from contaminating disulfide-linked aggregates. The isolated monomeric E2e reacts with a number of conformation-sensitive monoclonal antibodies, binds the soluble CD81 large external loop and efficiently inhibits infection of Huh7.5 cells by infectious HCV particles (HCVcc) in a dose-dependent manner, suggesting that it adopts a native conformation.
We determined the connectivity of the 9 disulfide bonds formed by 18 cysteines within the ectodomain of HCV glycoprotein E2, which are strictly conserved across HCV genotypes. Furthermore, circular dichroism combined with infrared spectroscopy analyses revealed the secondary structure contents of E2e, indicating in particular about 28% beta-sheet, in agreement with the consensus secondary structure predictions. The disulfide connectivity pattern, together with data on the CD81 binding site and reported E2 deletion mutants, enabled the threading of the E2e polypeptide chain onto the structural template of class II fusion proteins of related flavi- and alphaviruses. The resulting model of the tertiary organization of E2 shows the amino acid distribution among the characteristic class II domains, places the CD81 binding site at the interface of domains I and III, and highlights the location of a candidate fusion loop.
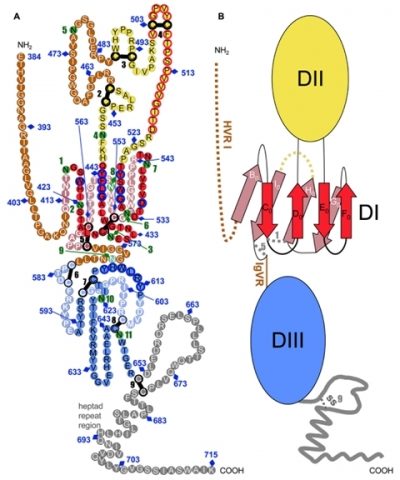
Tertiary organization of HCV E2e.
A) Disulfide bonds and glycosylation sites are indicated by thick black bars and green circles, respectively. Potentially unstructured segments are in brown, the stem region is grey. Residues that form the putative fusion loop region are encircled in red.
B) Schematic diagram of the tertiary organization of HCV E2, with DI, DII and DIII in red, yellow and blue, respectively. The β-strands in DI are labeled with the standard class II nomenclature.
Krey T, d’Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. (2010) PLoS Pathog 6: e1000762.
This work was supported by ANRS, Institut Pasteur, CNRS, the European Union, TOTAL and Merck-Serono. T.K. benefited from an Institut Pasteur ‘‘Bourse Roux’’ and then from an EU Marie Curie fellowship (MEIF-CT-2007-04725). C.M.K benefited from an ANRS post-doctoral fellowship. R.G.T. is funded by the E.U. HCVEI NTIR consortium.

