Présentation
Single cell phenotyping (flow cytometry) and cell sorting are supported by the Cytometry platform of the CB UTechS . The CB UTechS gathers expertise and high-end instrumentation for single cell phenotyping and sorting, single cell transcriptomics and single cell data analysis. We offer an end-to-end solution from study design, cell enrichment by FACS, single cell isolation and library preparation to bioinformatic data analysis. The team of dedicated professionals provides expert advice on experimental design, instrument training and support to users.
Multidimensional single cell profiling by conventional, spectral and imaging cytometry. Today’s flow cytometers enable an indepth characterization and quantification of protein expression patterns on live cells. State-of-the-art instruments available at the CB UTechS flow cytometry present multiplex advantages in single cell analysis, spanning from fast acquisition rate, large capacity of multiplexing (up to 30 parameters), management of autofluorescence (spectral cytometry), to spatial information in analysis of individual cells in suspension (imaging cytometry).
Single cell sorting. The cells of interest can be recovered by cell sorting in bulk or in single cell mode (in 96- or 384-well plates).
Support in single cell phenotyping. In addition to cutting-edge instrumentation, the CB UTechS proposes expertise in panel design, in establishing protocols for sample processing and preparation, in data collection and analysis (software Flow Jo, Kaluza, personalized non-supervised and supervised data analysis pipelines). Applications such as rare cells detection, micro-environment cytometry, apoptosis, cell cycle, cell activation, phospho-flow, and others are supported.
Technologies
 Conventional Cytometry. The main advantage of modern instruments is their capacity to simultaneously detect multiple fluorescently labeled cellular markers and thus to enable identification and characterization of various rare cell subsets from small sample volumes. In this respect, the most advanced technology is the FACSymphony™ (Becton Dickinson), capable of simultaneously detecting up to 30 parameters. Our platform is also equipped by a BD LSR Fortessa/ BD LSR II (5 lasers, 18 parameters) and a Beckman Coulter CytoFlex S (4 lasers, 13 parameters, plate reader) analyzer.
Conventional Cytometry. The main advantage of modern instruments is their capacity to simultaneously detect multiple fluorescently labeled cellular markers and thus to enable identification and characterization of various rare cell subsets from small sample volumes. In this respect, the most advanced technology is the FACSymphony™ (Becton Dickinson), capable of simultaneously detecting up to 30 parameters. Our platform is also equipped by a BD LSR Fortessa/ BD LSR II (5 lasers, 18 parameters) and a Beckman Coulter CytoFlex S (4 lasers, 13 parameters, plate reader) analyzer.
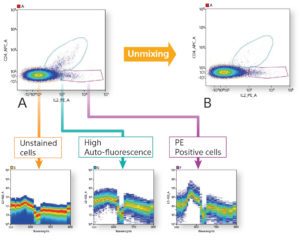 Spectral Cytometry. The spectral flow cytometry has revolutionarized flow cytometry by replacing the “one detector, one marker” paradigm by detection of full fluorescent signature of each dye. The SP6800 (Sony), equipped with three lasers, employs a prism array to disperse the collected fluorescent light over a 32‐channel multianode PMT. The measured spectra from individual cells are subsequently unmixed to allow increased precision in signal detection, lower background, higher sensitivity and a reliable identification of cell autofluorescence. The technology does not require compensation of unspecific signals and thus allows detection of highly ovelapping emission spectra and a flexibility in panel design .
Spectral Cytometry. The spectral flow cytometry has revolutionarized flow cytometry by replacing the “one detector, one marker” paradigm by detection of full fluorescent signature of each dye. The SP6800 (Sony), equipped with three lasers, employs a prism array to disperse the collected fluorescent light over a 32‐channel multianode PMT. The measured spectra from individual cells are subsequently unmixed to allow increased precision in signal detection, lower background, higher sensitivity and a reliable identification of cell autofluorescence. The technology does not require compensation of unspecific signals and thus allows detection of highly ovelapping emission spectra and a flexibility in panel design .
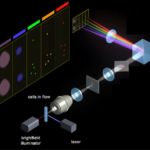 Imaging Cytometry. MARK II (Amnis, Luminex) integrates advantages of flow cytometry (5 lasers) and of microscopy (60x magnification) to simultaneously produce 12 images of each cell directly in flow, at rates exceeding 15 000 cells/min. It enables quantification of various intra- and inter-cellular processes in cells in suspension. Applications such as cell shape changes, internalization, apoptosis, nuclear translocation, co-localization, cell-cell interactions, spot-counting, cell cycle, cell senescence and many more are supported.
Imaging Cytometry. MARK II (Amnis, Luminex) integrates advantages of flow cytometry (5 lasers) and of microscopy (60x magnification) to simultaneously produce 12 images of each cell directly in flow, at rates exceeding 15 000 cells/min. It enables quantification of various intra- and inter-cellular processes in cells in suspension. Applications such as cell shape changes, internalization, apoptosis, nuclear translocation, co-localization, cell-cell interactions, spot-counting, cell cycle, cell senescence and many more are supported.
 Cell Sorting. Square cuvette and jet in air cell sorting instrumentation is available in BSL1, BSL2 and BLS2+ environments. We propose access to 3 BD FACS Aria III, 2 BD FACS ARIA Fusion and 2 MoFlo Astrios cell sorters. Cell sorting is usually the first step of different single cell analysis pipelines: sorting in bulk for 10xGenomics and similar technologies; single-cell sorting in 96-well plates for single cell real-time PCR; single cell sorting in 384 wells for MARS-Seq or SMART-Seq pipelines.
Cell Sorting. Square cuvette and jet in air cell sorting instrumentation is available in BSL1, BSL2 and BLS2+ environments. We propose access to 3 BD FACS Aria III, 2 BD FACS ARIA Fusion and 2 MoFlo Astrios cell sorters. Cell sorting is usually the first step of different single cell analysis pipelines: sorting in bulk for 10xGenomics and similar technologies; single-cell sorting in 96-well plates for single cell real-time PCR; single cell sorting in 384 wells for MARS-Seq or SMART-Seq pipelines.