About
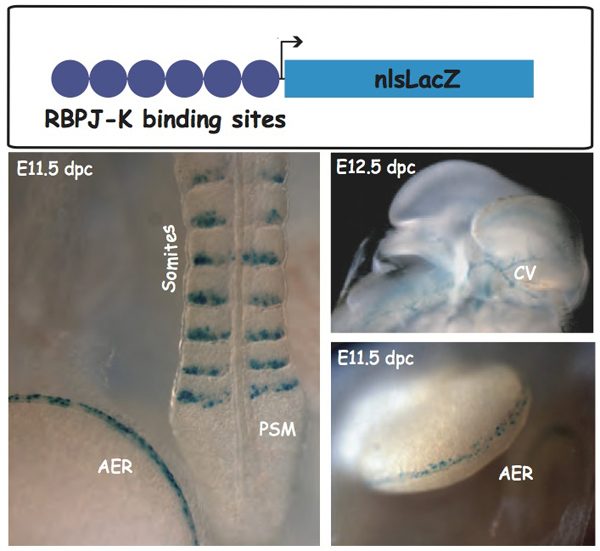
Because of the various levels of regulation of the Notch pathway, its activity cannot be easily deduced from the expression profiles of its components. In order to visualize Notch activity in situ at the cellular level, we produced a Notch reporter transgenic line that we named NAS for Notch Activity Sensor. We showed that transgene activation was coincident with known sites of activity of the Notch pathway and was abolished in an RBP‑Jk-deficient background, therefore indicating that the NAS mice allow the readout of the activity of RBP‑Jk/ Notch signaling pathway in vivo.
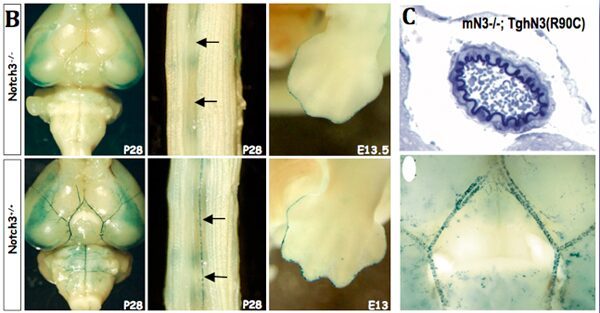
As other Notch reporter lines, NAS does not report on all Notch activity. In collaboration with Anne Joutel (INSERM U470, Paris), we have shown that it is particularly useful to monitor NOTCH3 activity. In Notch3-deficient mice transgene expression is abolished in arteries, while it is maintained at other sites therefore demonstrating that signaling through Notch3 receptor in arterial smooth muscle cells occurs through a RBP‑Jk-dependent pathway, although not involving the Hes/Hrt genes.
In addition, the NAS line has been very useful to bring new insights into the human vasculopathy CADASIL. CADASIL is an autosomal dominant small-vessel-disease of the brain caused by mutations in the Notch3 receptor. Mice expressing the human Notch3 R90C CADASIL mutation do not exhibit impaired receptor function, while mice expressing another mutation (C428S), that causes similar clinical features in humans, do. These results suggest that the pathogenesis of CADASIL disease does not primarily result from aberrant activity of the Notch3 signaling pathway.
The NAS line is available from the European Mouse Mutant Archive (EMMA-02412_strain_desc).
Publication related to NAS transgenic mouse line

Monet, M., Domenga, V., Lemaire, B., Souilhol, C., Langa, F., Babinet, C., Gridley, T., Tournier-Lasserve, E., Cohen-Tannoudji, M., and Joutel, A. (2007). The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum. Mol. Genet. 16, 982–992.
Monet-Leprêtre, M., Bardot, B., Lemaire, B., Domenga, V., Godin, O., Dichgans, M., Tournier-Lasserve, E., Cohen-Tannoudji, M., Chabriat, H., and Joutel, A. (2009). Distinct phenotypic and functional features of CADASIL mutations in the Notch3 ligand binding domain. Brain 132, 1601–1612.
Jory, A., Le Roux, I., Gayraud-Morel, B., Rocheteau, P., Cohen-Tannoudji, M., Cumano, A., and Tajbakhsh, S. (2009). Numb promotes an increase in skeletal muscle progenitor cells in the embryonic somite. Stem Cells 27, 2769–2780.
Publication related to other Notch reporter lines
Ohtsuka, T., Imayoshi, I., Shimojo, H., Nishi, E., Kageyama, R., and McConnell, S.K. (2006). Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Molecular and Cellular Neuroscience 31, 109–122.
Mizutani, K.-I., Yoon, K., Dang, L., Tokunaga, A., and Gaiano, N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355.
Vooijs, M., Ong, C.T., Hadland, B., HUPPERT, S., Liu, Z., Korving, J., van den Born, M., Stappenbeck, T., Wu, Y., Clevers, H., et al. (2006). Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development 134, 535–544.
Fre, S., Hannezo, E., Sale, S., Huyghe, M., Lafkas, D., Kissel, H., Louvi, A., Greve, J., Louvard, D., and Artavanis-Tsakonas, S. (2011). Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 6, e25785.
Smith, E., Claudinot, S., Lehal, R., Pellegrinet, L., Barrandon, Y., and Radtke, F. (2012). Generation and characterization of a Notch1 signaling-specific reporter mouse line. Genesis 50, 700–710.
Nowotschin, S., Xenopoulos, P., Schrode, N., and Hadjantonakis, A.-K.K. (2013). A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Developmental Biology 13, 15.
Liu, Z., Brunskill, E., Boyle, S., Chen, S., Turkoz, M., Guo, Y., Grant, R., and Kopan, R. (2015). Second-generation Notch1 activity-trap mouse line (N1IP::CreHI) provides a more comprehensive map of cells experiencing Notch1 activity. Development.