About
Perpetuating stemness
Neural stem cell (NSC) populations in the vertebrate brain generate adult-born neurons for plasticity, growth, and repair. Neurogenic and gliogenic capacity, based on long-term NSC maintenance, functionally define “stemness”. Stemness embodies massive NSC heterogeneity at the single cell level and requires control of maintenance or differentiation decisions at the population level.
PEPS is a multi-dimensional project exploring how spatiotemporal interactions among heterogeneous NSCs are coordinated to control population behaviour. It brings together experimental and theoretical groups with complementary expertise in NSC biology, biostatistics and mathematical modelling to decode the mechanistic principles of stemness.
Together, PEPS will lay the conceptual and methodological foundation to manipulate stem cell systems to improve their stability or output, and also produce new methods of universal value for studying cellular systems.
Project description
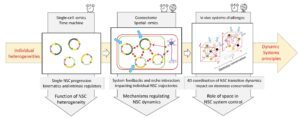
In an iterative experimental-mathematical approach, we will (1) solve the topology of individual NSC trajectories in transcriptomic space, (2) identify local cell-cell coordination mechanisms that impact these trajectories in situ, and (3) decode the resulting systemic properties and outputs of NSC ensembles at long-term and large spatial scales.
Aim 1: Decoding kinematics of individual NSC progression through the sc-omics feature space – Goal: We will combine experimental, modelling and in vivo validation steps to identify and describe the system kinematics, i.e., the distribution of individual NSC trajectories in the sc-transcriptomics feature space.
Aim 2- Characterising system-intrinsic feedbacks and external niche interactions modulating individual NSC trajectories – Goal. To decipher the network of interactions controlling trajectory dynamics, we will generate sc transcriptomic datasets reading cell identities and states in the neighbourhood of individual NSCs at any time, considering other system cells and external niche cells. These datasets will be statistically analysed to identify candidate regulatory feedbacks. They will be then iteratively tested by mathematical models and experimentally in vivo, proceeding in parallel in the vSVZ and ZfP and including young, intermediate, and aged adults.
Aim 3- Exploring the spatio-temporal coordination of structured NSC ensembles – Goal. We will generate static and dynamic datasets to describe the relative position of NSC trajectories in the physical space of the niche and whether and how this spatial distribution is restored after transient or local system perturbations. In parallel, we will apply different mathematical modelling strategies to resolve the structured NSC population dynamics in a spatially heterogeneous context, resolving the impact of the spatial scales of feedback signalling.
Methods
The PEPS project will use state-of-the-art methods in single cell -omics, lineage tracking, functional assays and quantitative image analysis in NSC models, analysed and integrated using through the development of novel biostatistics and modelling approaches.
We will focus on two biological model systems: the adult mouse ventricular sub-ventricular zone and zebrafish pallium, where NSC ensembles display comparable heterogeneity but differ in spatial organisation and fate dynamics.
This programme will result in original methods, including retrospective transcriptomics in single cells, innovative barcode transfers, and a novel mathematical framework to describe structured spatio-temporal population dynamics.
Partners
Laure Bally-Cuif
CNRS and Institut Pasteur, Paris, France
https://research.pasteur.fr/en/team/zebrafish-neurogenetics/
Ana Martin-Villalba
German Cancer Research Center (DKFZ), Heidelberg, Germany
https://martin-villalba-lab.github.io/
Simon Anders
Bioquant, Heidelberg University, Heidelberg, Germany
https://www.zmbh.uni-heidelberg.de/Anders/
Anna Marciniak-Czochra
Institute for Applied Mathematics, Heidelberg University, Heidelberg, Germany
http://www.biostruct.uni-hd.de/
Job opportunities
One postdoctoral position is available in the Bally-Cuif lab to decipher the transcriptomic trajectories of individual neural stem cells and their spatiotemporal regulation in vivo. The model system will be the neural stem cell niche of the zebrafish adult telencephalon. The project will combine sc-NMTseq, trajectory predictions in single cells, genetic approaches, intravital imaging and cell tracing in the adult animal, spatial neighborhood mapping in situ and quantitative image analysis.
Candidates must hold a Ph.D. in biology, have a strong interest in stem cell biology and/or zebrafish genetics, and a strong molecular background with preferably experience in single-cell -omics analyses. Proficiency in English is required.
More information at:
To apply, please submit a CV, motivation letter and at least two reference letters jointly to: