Project Description:
The field of connectomics has advanced significantly, allowing us to map synaptic connectivity of neural circuits at cellular resolution using electron microscopy. This internship project aims to develop and apply a novel class of models for analysing connectomic data, focusing on uncovering continuous hidden structures within these neural networks.
Objectives:
- Model Development: Extend traditional latent space models to analyse connectomic data, focusing on continuous latent variables representing neural tuning properties and connectivity functions.
- Data Analysis: Apply the developed models to synthetic connectomes and real-world datasets, such as the mouse retinal connectome, to uncover underlying structures.
- Validation: Assess the model’s performance in recovering known neural connectivity patterns and evaluate its scalability and flexibility in handling both directed and undirected graphs.
Methodology:
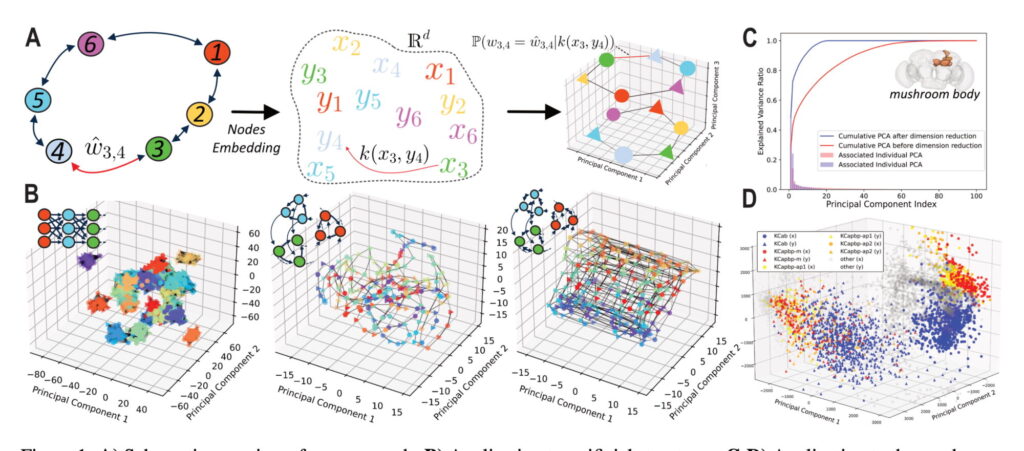
- Construct latent space graph models to estimate the probability of neural connections based on positions in a low-dimensional latent space.
- To fit the models, utilize advanced optimization techniques, such as quasi-Newton methods and stochastic gradient descent.
- Validate the approach by recovering classical connection topologies, such as ring attractors and synfire chains, from connectivity matrices.
Expected Outcomes:
- A robust model capable of revealing hidden structures in neural connectivity data.
- Insights into the organisational principles of neural circuits, contributing to our understanding of neural computation.
- Potential applications in neuroscience research, aiding in the interpretation of complex connectomic datasets.
Requirements:
- Strong background in statistical modeling, machine learning, or related fields.
- Proficiency in programming, particularly with Python.
- Familiarity with neural data analysis and connectomics is a plus.
Duration:
- The internship is expected to last for 6 months
Application:
Interested candidates should submit their CV, a cover letter, and any relevant academic transcripts to dbc-epi-recrutement at pasteur dot fr