Présentation
Early steps of infection
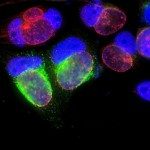
We have observed that chlamydia invasion of a host cell triggers a transient ubiquitination at the entry sites. Deubiquitination follows very rapidly (20 min post infection). We have identified a novel bacterial protein with deubiquitinase activity that likely participates in ubiquitin clearance early in infection. This deubiquitinase, which we named ChlaOTU, is shared by many chlamydial strains, including the human pathogen C. pneumoniae, but is absent from C. trachomatis. It possesses an amino-terminal signal for recognition by the type III secretion machinery. In addition to ubiquitin, ChlaOTU binds NDP52, a protein known for linking ubiquitinated substrates to autophagy compartments. One hypothesis is that ChlaOTU deubiquitinase activity helps Chlamydia escape recognition by the autophagy machinery of the host (Furtado 2013).
Identification of type III secreted proteins and functional studies
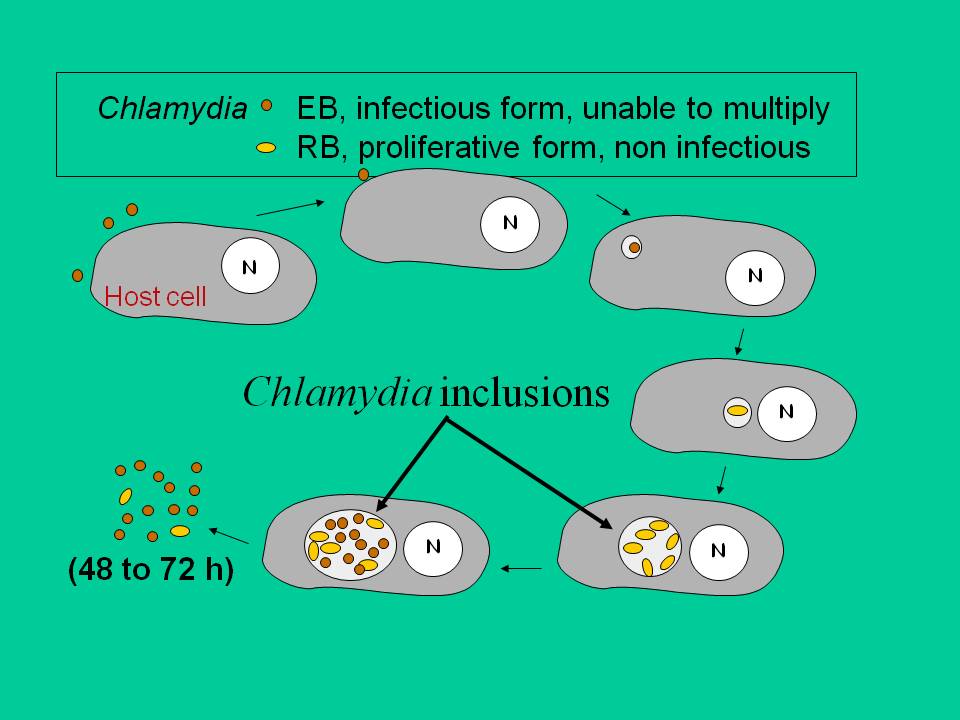
Throughout their cycle in the host cell, chlamydiae remain in a compartment called an inclusion. They use a type III secretion mechanism to translocate proteins into the host cell. These proteins are very likely to be important in Chlamydia pathogenicity and most of our research is dedicated to understanding their function during infection. We have shown that 7 to 10% of each chlamydial genome codes for proteins that remain anchored to the inclusion, where they probably govern transport to and across the inclusion membrane (Dehoux et al, 2011). We have identified a novel family of secreted proteins, with a common domain of unknown function (Muschiol et al, 2011). The expansion of this family of proteins in pathogenic chlamydiae and their conservation among the different species suggest that they play important roles in the infectious cycle, currently under study.
Living on host resources
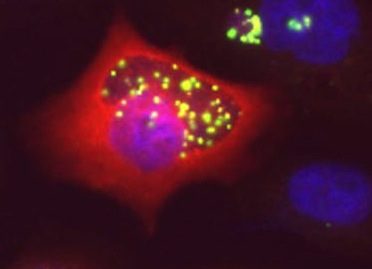
Chlamydiae uses host metabolites for growth. They lack enzymes for several biosynthetic pathways, including those to make some phospholipids, and exploit their host to compensate. Three-dimensional fluorescence microscopy demonstrates that small organelles of the host, peroxisomes, are translocated into the Chlamydia inclusion and are found adjacent to the bacteria. In cells deficient for peroxisome biogenesis the bacteria are able to multiply and give rise to infectious progeny, demonstrating that peroxisomes are not essential for bacterial development in vitro. Mass spectrometry-based lipidomics reveal the presence in C. trachomatis of plasmalogens, ether phospholipids whose synthesis begins in peroxisomes and have never been described in aerobic bacteria before. Some of the bacterial plasmalogens are novel structures containing bacteria-specific odd-chain fatty acids; they are not made in uninfected cells nor in peroxisome-deficient cells. Their biosynthesis is thus accomplished by the metabolic collaboration of peroxisomes and bacteria (Boncompain PLoS One 2014).