Présentation
Herpesviruses envelop glycoproteins
Herpesviridae are a large family of double-stranded DNA viruses, counting more than 200 members that infect humans and a wide range of invertebrates and vertebrates. Herpesviruses carry a large linear DNA genome of 100 – 200 Kb, which is packed in an icosahedral capsid. The nucleocapsid is immersed in a protein rich matrix called tegument, which is enveloped by a lipid bilayer decorated with a dozen of surface glycoproteins involved in virus attachment and entry.
Herpesviruses enter cells by fusion of the viral and plasma membranes. Membrane fusion is triggered by binding of a viral envelope protein to a specific cellular receptor, and is catalyzed by a three-part fusion machinery composed of the envelope glycoproteins B (gB) and hetero-dimeric non-covalent complex made of glycoproteins H and L (gH/gL). The fusion apparatus of herpesviruses (gB-gH/gL) is conserved.
We are interested in understanding the molecular mechanism of herpesvirus entry -how does the binding to a receptor activate the fusion machinery, and how does a complex of gB and gH/gL mediate the merger of lipid bilayers. We are using a combination of biochemical, biophysical and structural biology methods to address these questions. One of the main goals is to determine three dimensional structures of the proteins involved in the entry process by X-ray crystallography. We are currently expressing a range of recombinant proteins from alpha-herpesviruses Herpes simplex virus 1, Varicella zoster virus and Pseudorabies virus (PrV).
We have recently solved the structure of a core fragment of PrV gH (gHC) in complex with an antibody. PrV gHC lacks an N-terminal domain, does not associate with gL, and resembles a functional, naturally occurring gH truncation, which is fused to gD, the receptor binding protein, and is found in a gL-null virus (Klupp BG, Mettenleiter TC, J Virol, 1999). This gDgH chimera mediates gL-independent fusion, and gHC therefore represents the core gH domain sufficient to carry out fusion.
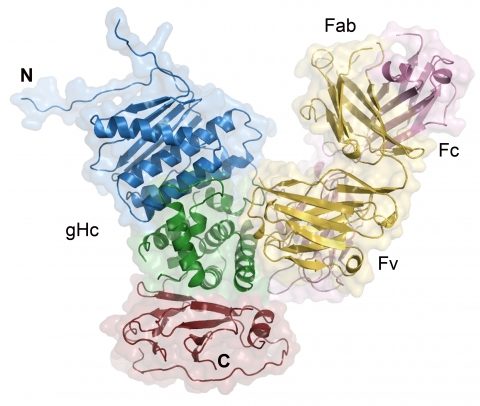
Structure of the PrV gHC:Fab d6.3 complex.
PrV gHC is colored in blue, green and red, blue corresponding to the N- and red to the C-terminus. Heavy and light chains of the Fab are colored in light magenta and yellow, respectively.
Structure of a core fragment of glycoprotein H from Pseudorabies virus in complex with antibody. Backovic M, Dubois R, Cockburn JJ, Sharff AJ, Vaney MC, Granzow H, Klupp BG, Bricogne G, Mettenleiter TC, Rey FA. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22635-40.
This work was supported by the French ‘Ministère de l’Economie, de l’Industrie et de l’Emploi’ (CristaLead grant), the French ‘Agence Nationale de Recherche’ (ANR, DENtry, program ‘Microbiologie, Infections et Immunité’), by Merck-Serono and by recurrent fundings from Institut Pasteur and CNRS.