Présentation
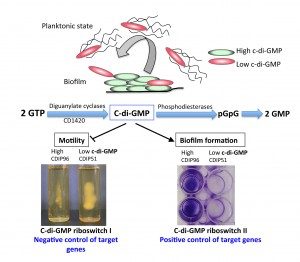
In bacteria, it is increasingly recognized that ncRNAs play a critical role in adaptive responses and in various metabolic, physiological and pathogenic processes. In Clostridium difficile, by combining in silico analysis, RNAseq and genome-wide promoter mapping, we identified more than 200 ncRNAs (Soutourina et al, PLos Genetics, 2013). This includes trans riboregulators located in intergenic regions, cis-antisense RNAs, riboswitches (5’-cis-regulatory elements) and CRISPR (clustered regularly interspaced short palindromic repeats) RNAs. Since the mechanisms controlling Clostridium difficile infection cycle are complex, the high number of ncRNAs in this pathogen strongly suggested their importance in the control of gene expression. Among the ncRNA identified, we are particularly interested in the riboswitches responding to an important signaling molecule: cyclic-di-guanosyl-5’monophosphate (c-di-GMP). C-di-GMP controls lifestyle switch from free-living motile state to biofilm communities and virulence in bacteria. In contrast to most Gram-positive bacteria, Clostridium difficile encodes a large number of c-di-GMP turnover enzymes (up to 37) suggesting an important role for this molecule. The originality of c-di-GMP-dependent control in Clostridium difficile is the use of RNA molecules as effectors instead of protein effectors found in the majority of ba