Présentation
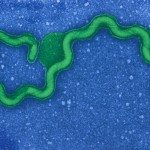
Leptospira spp. are thin, highly motile, slow-growing spirochetes that can be distinguished from other bacteria on the basis of their unique helical shape and the presence of periplasmic flagella.
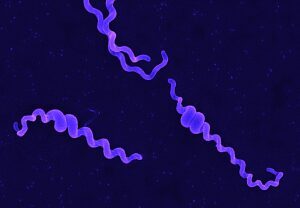
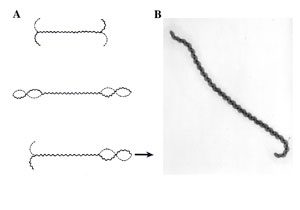
Movement of leptospires (A, from Goldstein and Charon, 1988) and electronic microscopy of Leptospira biflexa (B). During translational motility, counterclockwise rotation of the perisplasmic flagellum produces spiral-shaped ends at the leading edge while concomitantly, clockwise PF rotation causes gyrating cell ends at the tail to remain hooked-shaped
The basal body and hook structures of spirochete flagella are similar to those of the model systems of Escherichia coli and Salmonella enterica. However, in contrast to the flagellin polymer of enterobacteria, the flagellar filaments of spirochetes investigated to date have a complex structure comprising FlaA and FlaB proteins. In Leptospira spp. the flaA genes are organized in a locus with flaA2 immediately upstream of flaA1. We showed that the flaA1 and flaA2 transposon mutants were able to assemble periplasmic flagella, but their motility, virulence, and flagellar and cellular morphologies were affected, depending on whether FlaA1 or both FlaA proteins were expressed. Our most recent studies using whole genome sequencing of spontaneous non-motile strains and preliminary screening of a library of transposon mutants for motility-deficient mutants allowed the identification of two novel flagellar proteins named FcpA and FcpB