Présentation
Centre for the Analysis of Complex Systems in Complex Environments
About CACSICE
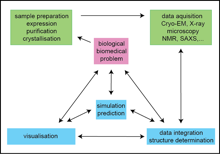
Cells function through transient interactions of macromolecular complexes. Understanding fully the dynamic processes of life at the molecular level requies understanding the structures of these dynamic complexes. Developing the tools to accomplish this aim is the subject of this proposal: we will create a centre dedicated to studying the spatio-temporal complexity of macromolecular assemblies and macromolecules in living systems. Historically, structural biology has been largely confined to studying static structures of a single molecule with a single technique, usually X-ray crystallography. To understand the formation and evolution of transient complexes within a living cell, we need a radical change in structural biology approaches. Data acquired with multiple biophysical techniques at multiple scales must be collected and integrated into one consistent, dynamic, picture that relies both on emerging experimental technologies and on modelling and numerical simulations. The studied processes come from several areas of biology, such as biochemistry of membranes and nucleic acid systems, structural virology, microbiology, cellular biology, and neurobiology. Studying these complex structures on multiple time scales requires many biophysical techniques, including ultra resolution light and electron microscopy, X-ray crystallography, liquid and solid state NMR, small angle X-ray scattering and structural mass spectrometry. Data analysis and multi-technique data integration, modelling approaches for dynamics and protein-ligand interactions, bioinformatics, and advanced visualisation of experimental and modelling results, will play a central role. The success of CACSICE will build on the internationally acknowledged scientific excellence of the participating groups and on the long-standing experience of its core facilities in providing expertise to the scientific community in Paris and beyond. CACSICE will be one of very few resource centres of its kind, permitting researchers to pursue multi-technique studies systematically and relieving them of the onerous need to collaborate with groups in other countries in order to achieve their aims, which is unacceptable given the leading technological status France. Frequent formal and informal meetings will address the scientific problems efficiently and encourage the development of new, ground-breaking methods. CACSICE will change the way that challenging problems in structural biology are addressed. Projects will be tightly integrated with the research of world-wide leading institutes in biochemical and biomedical research, stimulating numerous new research projects with increasing complexity and biological relevance and closing the resolution gap between structural and cellular biology. This will not only lead to major discoveries through fundamental understanding of biological processes, but also produce technical benefits through the development of innovative ways to study biological structures. CACSICE will not only provide access to a complete set of state-of-the-art equipment but also provides a pool of expertise for other researchers in France and beyond, in both the academic and private sectors. The partners in this proposal already have numerous collaborations with each other, locally, nationally and internationally. CACSICE will foster new cross-discipline collaborations. The scientific projects and the tight integration of methodologies and of length time scales are at the forefront of structure biology and are innovative in the scope, integration and completeness of the approaches. Our work should identify new drug targets and therapeutic strategies. This proposal will structure the community of biophysical chemists around two major core facilities (Institut Pasteur and IBPC) and two neighbouring institutes that bring expertise and excellence in specific areas, providing a nexus for innovation.
Partners
- Institut Pasteur Bioinformatique structurale / CNRS UMR3528
- Institut Pasteur Virologie structurale / CNRS URA3015
- Institut Pasteur Résonance magnétique nucléaire des biomolécules / CNRS UMR3528
- Institut Pasteur Biologie structurale de la sécrétion bactérienne / CNRS UMR3528
- Institut Pasteur Biochimie des Interactions macromoléculaires / CNRS UMR3528
- Institut Pasteur Imagerie et modélisation / CNRS URA2582
- Institut Pasteur Imagerie Dynamique du Neurone / CNRS URA2182
- Institut Pasteur Neuroscience moléculaire et transport membranaire / CNRS URA2182
- Institut Pasteur Génétique moléculaire / CNRS ERL3526
- Institut Pasteur Proteopole / CNRS UMR3528
- Institut Pasteur Dynamique structurale des macromolécules / CNRS UMR3528
- Institut Pasteur Récepteurs-canaux / CNRS URA2182
- Institut Pasteur Spectrométrie de Masse Structurale et Protéomique / CNRS UMR3528
- CNRS UMR7099 Laboratoire de Biologie Physico-Chimique des Protéines membranaires / UP Diderot
- CNRS UPR9080 Laboratoire de Biochimie Théorique / UP Diderot
- CNRS FRE3354 Laboratoire de Biologie Moléculaire et Cellulaire des Eucaryotes / UPMC
- CNRS UPR9073 Expression Génétique Microbienne
- CNRS UMR7141 Physiologie membranaire et moléculaire du chloroplaste / UPMC
- CNRS UMR8015 Laboratoire de cristallographie et RMN biologiques / UP Descartes
- CNRS UMR7590 Structure et Dynamique des Protéines / UPMC
- CNRS UMR8601 Equipe de RMN des Substances d’Intérêt Biologique / UP Descartes
- CNRS FRC550 Institut de Biologie Physico-Chimique
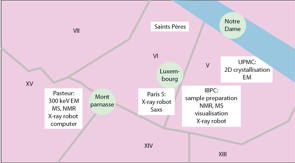
Equipment
Existing
- 200 kV electron microscope with Falcon II direct detection camera
- Synapt mass spectrometer for characterisation of native macromolecular complexes and HDX
Planned
- 300 kV electron microscope (IP)
- Mass spectrometer for post-translational modifications (IBPC)
- 700 MHz NMR spectrometer for solid and liquid NMR (IBPC)
- NMR with F19 probe (Paris Descartes)
- SAXS (Paris Descartes)
- 2D crystallisation robot (UPMC)
- Visualisation wall (IBPC)
- Computing cluster (IP)


