Research Engineer/Postdoctoral Position
Decision and Bayesian Computation (DBC) – Epiméthée (EPI) Laboratory
Institut Pasteur, Paris | 25 rue du Docteur Roux, 75015 Paris
Position Overview
We are seeking two motivated researchers or engineers with expertise in finite element programming to join an ambitious project modelling the complete neuromuscular dynamics of Drosophila larvae. This position offers a unique opportunity to work at the intersection of computational physics, neuroscience, and biomechanics, developing physics-based simulations that bridge neural circuits, muscle activity, and whole-body behaviour.
Start date: Flexible
Duration: 2 years (engineer positions can transition to postdoctoral positions)
Positions available: 2
The Project
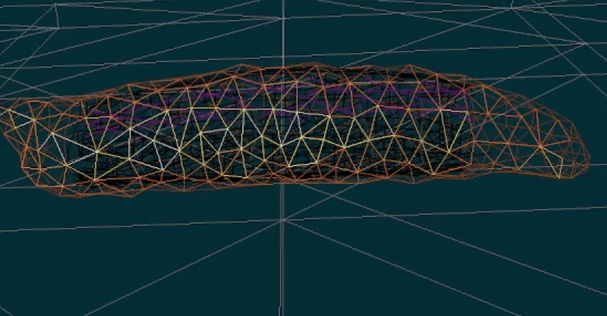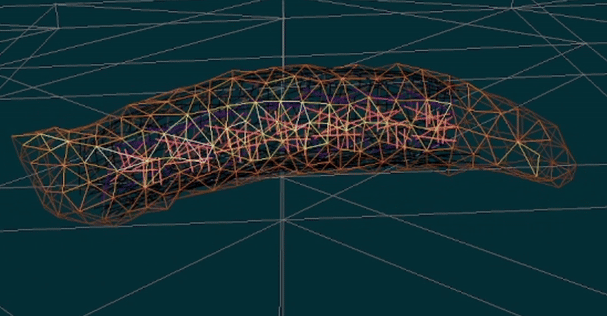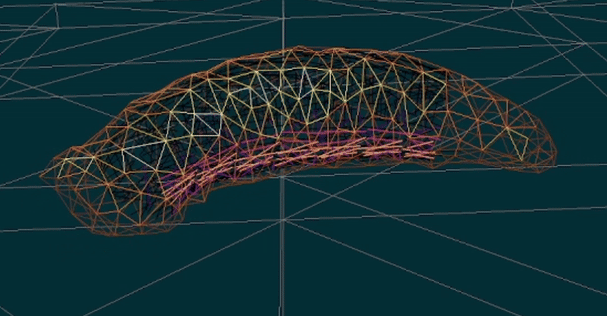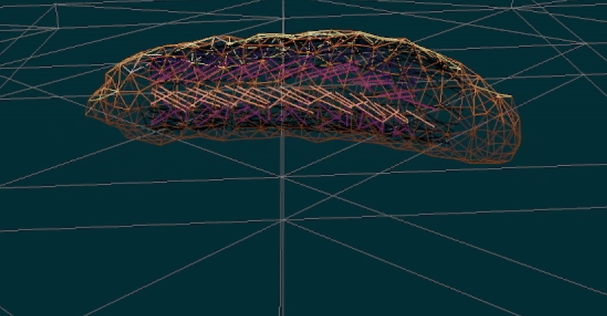
You will build upon and extend our existing functional simulation platform for Drosophila larva locomotion, which already incorporates muscle and body dynamics models. Your role will be to refine, validate, and expand this simulation framework to accurately replicate the larva’s complete neuromuscular control system—a highly deformable biological system with a fully mapped neural connectome.
The project leverages:
- Our current physics-based simulation with implemented muscle and body mechanics
- Recently acquired data on muscle activity patterns linked to specific behaviours
- Complete synaptic connectome of the larval nervous system
- High-resolution micro-CT scans of muscular structure
- Large-scale behavioural datasets
Core objectives:
- Refine and optimise the existing simulation framework to improve accuracy and performance
- Infer and constrain model parameters using experimental muscle and neural recordings
- Explore motor control policies that replicate observed behaviours
- Test simulation predictions against muscle ablation experiments
- Investigate how environmental physics affects larval actions
- Develop robust APIs for community access
Required Qualifications
Essential:
- Strong background in finite element methods and numerical simulation
- Proficiency in Python and/or C++
- Experience with physics-based modelling of deformable bodies or soft materials
- Solid understanding of computational mechanics and dynamics
Highly Desired:
- Direct experience with the SOFA framework (Simulation Open Framework Architecture)
- High-performance computing and cluster computing experience
- Background in biomechanics, robotics, or computational neuroscience
Additional Assets:
- Experience with machine learning approaches
- Knowledge of muscle mechanics (Hill muscle model or similar)
- Previous work on simulated bodies or animal locomotion
Your Role
You will work collaboratively with a multidisciplinary team to:
- Extend simulation frameworks: Build upon our existing larva simulation to incorporate more detailed muscular geometry, improved dynamics, and better integration with experimental data
- Optimise and validate: Constrain free parameters using experimental data from muscle calcium imaging and behavioural recordings.
- Explore motor control: Apply computational approaches (probabilistic programming, imitation learning) to reproduce and predict larval behaviours
- Deploy tools: Create accessible APIs enabling the broader scientific community to explore motor control across various computational settings
The Laboratory
The DBC-EPI laboratory is a mixed research unit (Institut Pasteur, INRIA, CNRS, UPC) focused on understanding the organizing principles of biological information processing through embodied neuroAI. Led by Jean-Baptiste Masson (DR, theoretical physicist), with deputies Christian Vestergaard (CR), Alex Barbier-Chebbah (CR), and François Laurent (IR, applied mathematician and software engineer), the lab combines theoretical physics, advanced statistics, and computational approaches to study neural circuits, decision-making, and sensory processing.
Recent achievements include developing platforms for medical imaging analysis (DIVA, now Avatar Medical—a successful spinoff), studying neural connectomes in Drosophila, and work in simulation-based inference for biological systems.
You will work directly with:
- François Laurent – Project coordinator, expert in computational frameworks and behavioral analysis
- Alex Barbier-Chebbah – Theoretical physicist
- Jean-Baptiste Masson – Lab director with expertise in biological information processing
Working Environment
- Modern computational infrastructure with access to high-performance clusters
- Collaborative environment at Institut Pasteur’s vibrant campus in central Paris
- International network including partnerships with Janelia Research Campus (HHMI) and leading European labs
- Active participation in seminars, workshops, and conferences
- Opportunities for both fundamental research and applied technology development
Salary
- The researcher or engineer with 1-3 years after PhD would be ~€48K gross/year at ~€3,120 net/month
- The researcher or engineer with 1-3 years after Master would be ~€45K gross/year at ~€2,970 net/month
- Salaries can be adjusted to the specificities of the candidate
Application Process
Please submit:
- CV with publication list (if applicable)
- Contact information for 1-2 references
Contact:
François Laurent & Jean-Baptiste Masson: dbc-epi-recrutement AT pasteur.fr
Visit our lab website for more information about our research: https://research.pasteur.fr/en/team/decision-and-bayesian-computation/
Institut Pasteur and INRIA are committed to diversity and equal opportunity employment. We welcome applications from candidates of all backgrounds.