Link to Pubmed [PMID] – 25629728
PLoS ONE 2015;10(1):e0117163
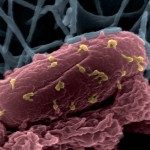
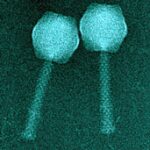
In a previous study, six virulent bacteriophages PAK_P1, PAK_P2, PAK_P3, PAK_P4, PAK_P5 and CHA_P1 were evaluated for their in vivo efficacy in treating Pseudomonas aeruginosa infections using a mouse model of lung infection. Here, we show that their genomes are closely related to five other Pseudomonas phages and allow a subdivision into two clades, PAK_P1-like and KPP10-like viruses, based on differences in genome size, %GC and genomic contents, as well as number of tRNAs. These two clades are well delineated, with a mean of 86% and 92% of proteins considered homologous within individual clades, and 25% proteins considered homologous between the two clades. By ESI-MS/MS analysis we determined that their virions are composed of at least 25 different proteins and electron microscopy revealed a morphology identical to the hallmark Salmonella phage Felix O1. A search for additional bacteriophage homologs, using profiles of protein families defined from the analysis of the 11 genomes, identified 10 additional candidates infecting hosts from different species. By carrying out a phylogenetic analysis using these 21 genomes we were able to define a new subfamily of viruses, the Felixounavirinae within the Myoviridae family. The new Felixounavirinae subfamily includes three genera: Felixounalikevirus, PAK_P1likevirus and KPP10likevirus. Sequencing genomes of bacteriophages with therapeutic potential increases the quantity of genomic data on closely related bacteriophages, leading to establishment of new taxonomic clades and the development of strategies for analyzing viral genomes as presented in this article.