About
Combining bioinformatics and immunobiology towards the identification of Trypanosoma vivax specific markers.
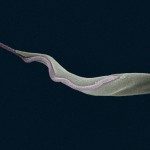
Despite closely related, South American and West African T. vivax are genetically distinct and these populations largely differ from those circulating in East Africa. The specific diagnosis of T. vivax is crucial to assess the real prevalence of this pathogen and the first step to better understand the population structure and any link between genotypes and differences in pathogenicity, virulence and drug resistance. However, parasitological methods are not sensitive, serological assays are not highly specific and both molecular and serological diagnoses of T. vivax are unable to detect all genotypes found across Africa and South America. Therefore, new methods for the diagnosis of T. vivax infections are urgently required and the identification of new candidates for both molecular and serological diagnoses is one of the main aims of this project.
The project intends to compare the available transcriptome from the African T. vivax strain IL1392 [1] , with the transcriptome of blood forms of one Brazilian isolate that we are planning to obtain within this study by NGS using the Illumina HiSeq platform. This analysis will be valuable to identify markers that can be useful for diagnosis and genotyping of T. vivax and, hence, to explore how genetic variation among isolates of T. vivax impacts on the spread of disease and pathogenicity.
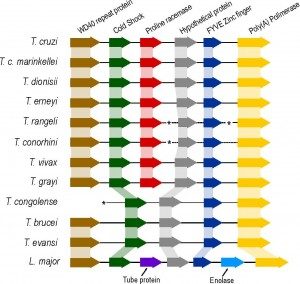
[1] A recent collaborative study of the Tip-IP laboratory with A.P. Jackson, Wellcome Trust Research Fellow at University of Liverpool, has generated 13 libraries corresponding to data sets for bloodstream, metacyclic and epimastigote forms of the parasite produced in Paris. RNAseq data in FASRQ format was made available for the strain IL1392 through the Sanger Institute FTP site.