About
Gene expression is controlled and regulated by nuclear and cytoplasmic surveillance mechanisms in order to preserve the integrity of genetic information. Some of the mechanisms occur in the nucleus during transcription and maturation of mRNAs; they allow recognition and degradation of non-conform mRNAs in the nucleus. Nevertheless, some aberrant mRNAs such as those lacking a Stop codon (Non-Stop mRNA) are exported to the cytoplasm. Because they contain a 5’ cap and a 3’ polyA tail, they are recognized by the translational machinery, engage in translation, but are unlikely to terminate translation properly. Other deleterious features found within mRNAs can interfer with proper translation and lead to stalling of ribosomes (No-Go mRNA). These events lead to elongated or truncated peptides, which may have toxic effect. Cytoplasmic quality control processes recognize ribosomes that are stalled while translating and selectively eliminate the aberrant mRNAs as well as the corresponding aberrant polypeptides to maintain protein homeostasis.
Factors responsible for non-conform mRNAs degradation are relatively well described and involve cytosolic RNA exonucleases in combination with factors that are specific to the different aberrant translation events. The studies of processes targeting nascent polypeptides issued from these aberrant RNAs is, however, much less advanced and, apart form an E3 ubiquitin ligase, Ltn1, described as a ribosome-associated protein involved in the ubiquitinylation of nascent non-conform polypeptides, the factors involved in degradation of the resulting aberrant proteins were unknown when we started this project.
The goal of my team project is to identify factors involved in peptides degradation generated from aberrant mRNA and to understand the mechanism of action of these factors.
In the current model for the Non-Stop mRNA decay, the ribosome translate the polyA tail in polylysine which result in ribosome stalling and translation arrest. Ribosome’s empty A site is recognized by Dom34 and Hbs1, the orthologs of eRF1 and eRF3 translation termination factors, except that Dom34/Hbs1 are unable to do the peptidyl-tRNA cleavage. After GTP hydrolysis by Hbs1, Rli1 is recruited leading to ribosomal subunits dissociation. In addition, the SKI complex, together with the exosome, is involved in the degradation of the aberrant mRNA (NSD and NGD pathway). Whereas the exosome can act either in the nuclear or cytoplasmic compartment, the SKI complex is strictly cytoplasmic.
To identify new partners involved in this translational quality control, and notably in the degradation of the aberrant peptides, we first performed genome-wide genetic screens.
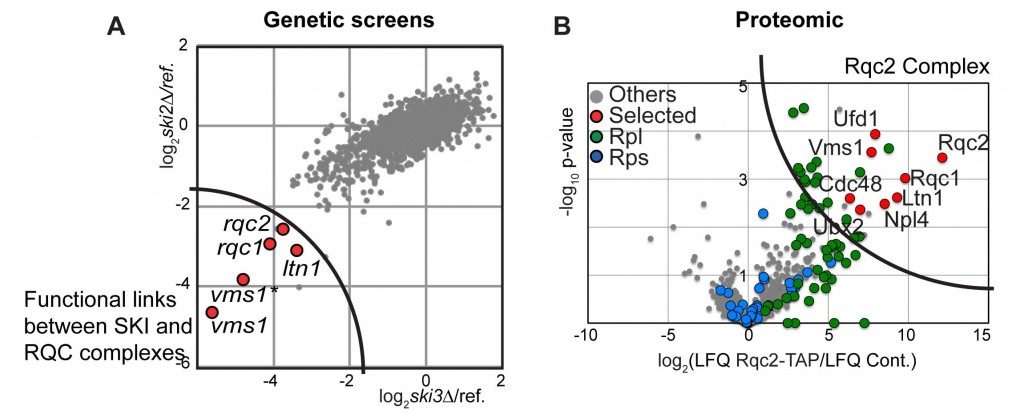
The deletion of the gene coding for the SKI complex is viable in classical growth culture conditions showing that the Non-Stop mRNA decay pathway is not essential. We postulated that mutations into factors involved in the degradation of aberrant peptides could be deleterious for the cells, when associated to SKI mutations, which stabilise the aberrant mRNAs. To address this possibility, we performed synthetic lethal screens using mutants of the SKI complex. We selected three factors, Ltn1, Ypl009c/Rqc2 and Ydr333c/Rqc1, which indeed became essential when the SKI complex was affected and the translational fidelity reduced.
By biochemical affinity purification using these factors as bait, we identified a new complex formed of Ltn1, Rqc2 and Rqc1 and the hexamer of ATPase Cdc48 now named the RQC complex for “Ribosome Quality Control”.
This complex is associated to the large ribosomal subunit containing the aberrant peptides blocked in the exit tunnel of the 60S particles. By affinity purification in several mutant conditions, together with functional tests, we observed that Rqc1 and Ltn1 are required for Cdc48’s binding (Defenouillère et al, PNAS 2013).
Recently, Shen et al., (Science 2015) demonstrated that Rqc2 is required for the addition of CAT tail of the aberrant nascent peptide and few years ago Bengtson and Joazairo (Nature 2010), showed that Ltn1 polyubiquitylated aberrant nascent peptides.
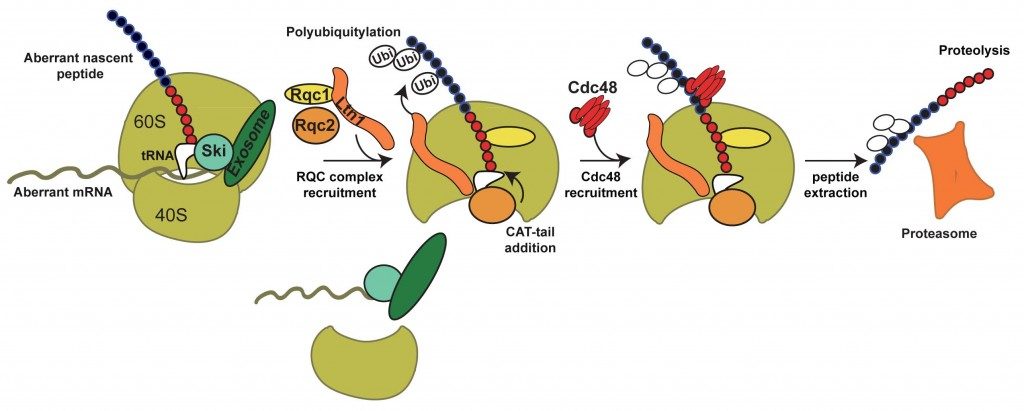
Altogether, these results allowed us to propose a model for the selective degradation of aberrant peptides generated from Non-Stop mRNAs.
In summary, the aberrant peptides generated from Non-Stop mRNA stalled on the 60S are recognized by the RQC complex. The polyubiquitylation of the aberrant peptide by Ltn1 and the presence of Rqc1 are required for the binding of Cdc48, which would extract the peptide from the exit tunnel. After, extraction, the peptide is degraded by the proteasome. We are now investigating the molecular role of these factors, notably Rqc1, and the mechanisms of recognition and degradation of the aberrant translation products.