About
Double-stranded (ds) RNA virus particles are organized around a central icosahedral core capsid made of 120 identical subunits. This core capsid is unable to invade cells from outside, and animal dsRNA viruses have acquired surrounding capsid layers that are used to deliver a transcriptionally active core particle across the membrane during cell entry. In contrast, dsRNA viruses infecting primitive eukaryotes have only a simple core capsid, and as a consequence are transmitted only vertically.
We solved the 3.4 Å X-ray structure of a picobirnavirus—an animal dsRNA virus associated with diarrhoea and gastroenteritis in humans. The structure shows a simple core capsid with a distinctive icosahedral arrangement, displaying 60 two-fold symmetric dimers of a coat protein (CP) with a new 3D-fold. We show that, like many non-enveloped animal viruses, CP undergoes an autoproteolytic cleavage, releasing a post-translationally modified peptide that remains associated with nucleic acid within the capsid. Our data also show that picobirnavirus particles are capable of disrupting biological membranes in vitro, indicating that its simple 120-subunits capsid has evolved animal cell invasion properties.
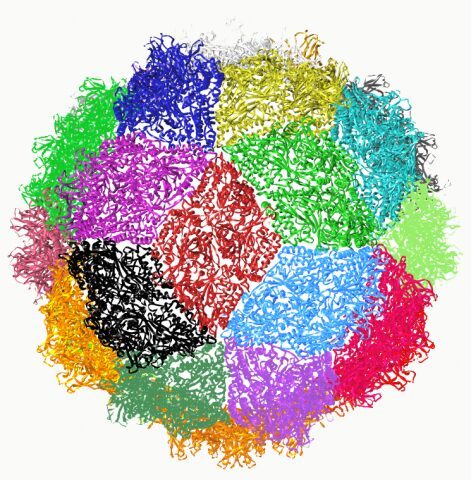
Structure of the icosaedral picornavirus particle.
A) Structure of ds RNA segment 1 from PBV.
B) The CP forms a perfectly symmetrical dimer related by a local two-fold axis L2.The N-terminal residues are exchanged between the two subunits generating an intricate interface.
C) The icosaedral two-fold axis of the particle relates two CP dimers such that they form a rhombus or diamond tile. This axis is perpendicular to the view at the intersection the 5- (I5) and 3-fold (I3) icosaedral axis.
D) Together, the 30 tiles generated by icosaedral two-fold axis give rise to a spherical particle with triacontahedral design.
The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA.EMBO J. 2009 Jun 3;28(11):1655-65.
This work was supported by CNRS, INRA, Institut Pasteur, Université Paris XI and Merck-Serono.

