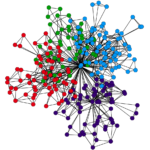
I’m a PhD student in statistical physics, I’m studying the representation of the Drosophila larva’s connectome as an ensemble of canonical microcircuits. The Drosophila larva is a great model animal for (micro-)connectomics, since its wiring diagram, at level of single synapses, has almost been completely mapped. The structure of the larva’s brain is far from random, as many other higher-species’ brains which are often modeled as small-world networks. The genome, as an information reservoir, is too small to encode such a complex structure. The genomic bottleneck principle actually states that the genome makes use of some encoding strategies to constrain the brain structure, which only means that the connectome is compressed within the DNA. In information theory, compression is an expression of regularities in a system. Thus, we consider, for the Drosophila larva’s connectome, canonical microcircuits as mesoscale structural regularities, or network motifs, supporting the hypothesis of a genomic compression. We explore this representation we use tools from network science and information theory to establish the statistical significance of those canonical microcircuits.
© Research