About
The identification of genetic markers of major family strains opened the way to the study of DNA repair in M. tuberculosis
In M. tuberculosis the acquisition of an antibiotic multiresistant phenotype is due to the accumulation of mutations in antibiotic targets and not to horizontal transfer as described for outbreaks of other antibiotic multiresistant bacteria of clinical importance. Because many of these targets correspond to essential pathway, we hypothesised that M. tuberculosis multidrug resistant (MDR) strains that are responsible for outbreaks might correspond to isolates that have acquired advantages and that are more adapted to their host. We began to investigate the role of DNA repair genes in the adaptation of M. tuberculosis to its host. The W-Beijing family is responsible for the majority of MDR TB cases in Eastern Europe. A first analysis of the polymorphism of DNA repair genes in this family had identified the presence of important variations in two putative mutT and one ogt gene.
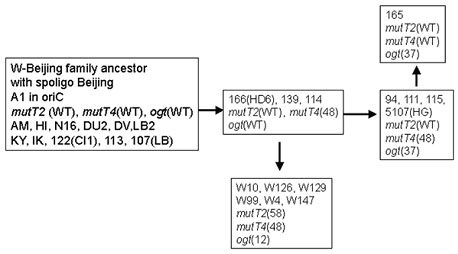
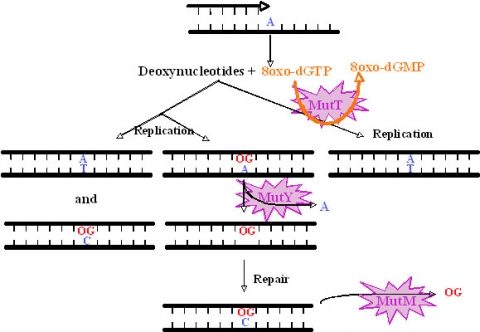
We have studied the DNA repair system that is responsible for the damage prevention and repair of one of the most frequently occurring stable and abundant oxidized base lesions in DNA, the 8-oxo-7,8-dihydroxy guanine (8-oxo-G). This system comprises the MutT, MutY and MutM enzymes and is commonly denominated the GO repair system. Four M. tuberculosis ORFs showing similarities with E.coli mutT were found. They were named mutT1 to T4. To create the mutants we used the ts/sacB allelic replacement system previously developed by our laboratory. The analysis of the mutants capacity to generate spontaneous mutations in an adapted Luria-Delbruck fluctuation test was performed. MutT1 deficiency in M. tuberculosis resulted in a 15.5-fold spontaneous mutation frequency increase by rifampicin resistance (RifR) screening compared with the wild type strain. A similar 12-fold increase was observed for the mutT1 mutant of M. smegmatis. Furthermore, we observed a striking 48.1-fold increase in spontaneous RifR colonies for the MutT4-deficient strain. One may hypothesize the existence of enzymes with functions redundant to that of MutT4 in M. tuberculosis or a different regulation of gene expression in the two species. Defects in mutT2 and mutT3 genes resulted in no apparent differences between mutant and wild type strains. For M. tuberculosis, with the exception of the dnaE2 role in inducible mutagenesis, no other gene was found to be associated with a mutator or antimutator phenotype.
All mutant phenotypes above described were abrogated by the complementation with wild type alleles. As suggested by mutations identified in rpoB during the fluctuation tests, activities of MuT1, Mut2 and MutT4 proteins have suggested a clear 8-oxoGTPase activity and a hydrolytic activity on dGTP (as described for E. coli MutT).
Polymorphism of 3R genes in M. tuberculosis isolates
We have analysed the polymorphism of 56 genes involved in DNA repair, replication and recombination (3R genes) in a representative collection of 92 clinical isolates. We identified essentially single nucleotide polymorphisms. A higher level of polymorphism was observed in 3R genes as compared to other house-keeping genes. We identified 192 polymorphisms, among them, 161 non synonymous (ns) and 91 synonymous (s) SNPs. We identified 74 haplotypes, with nucleotide diversity per site of 0.00024, approximatively twice that reported for the control group of housekeeping genes (3R genes being excluded from this group). Site frequency spectrum comparison of synonymous and non-synonymous variants and Ka/Ks ratio analysis suggest a general negative/purifying selection acting on these sets of genes that may lead to suboptimal 3R system activity. This situation, and the consequent lack of fidelity in genome maintenance, may serve as a starting point for the evolution of antibiotic resistance, fitness for survival and pathogenicity, possibly conferring a selective advantage in certain stressful situations. These findings suggest that 3R genes may play an important role in the evolution of highly clonal bacteria, such as M. tuberculosis. A 3R-based phylogenetic tree was constructed. This tree is congruent with one constructed with other markers like the DR repeats. It is a new tool for distinguishing between M. tuberculosis complex strains. With many more microbial genomes being sequenced, our results open the door to 3R gene-based studies of adaptation and evolution of other, highly clonal bacteria.
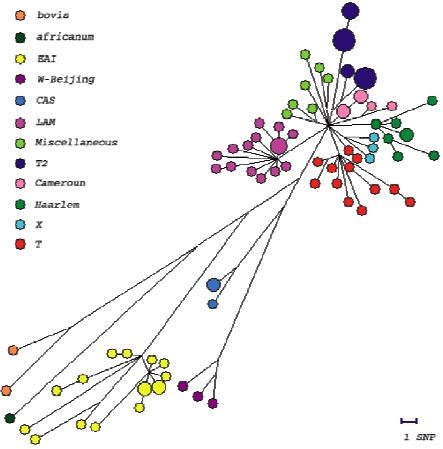
Among the SNPs above described, one of them (within mutT1) was used to show the predominance of a strain family in MDR isolates in CAR, thus identifying strains at higher risk to become MDR. Another SNP corresponding to a nonsense codon in the fused ada alkA gene in two distant family strains allowed to identify a case of convergent evolution. This may suggest an association of this SNP to a selective advantage.
