About
Integration of single-cell multi-omics data across space and time to unlock cellular trajectories
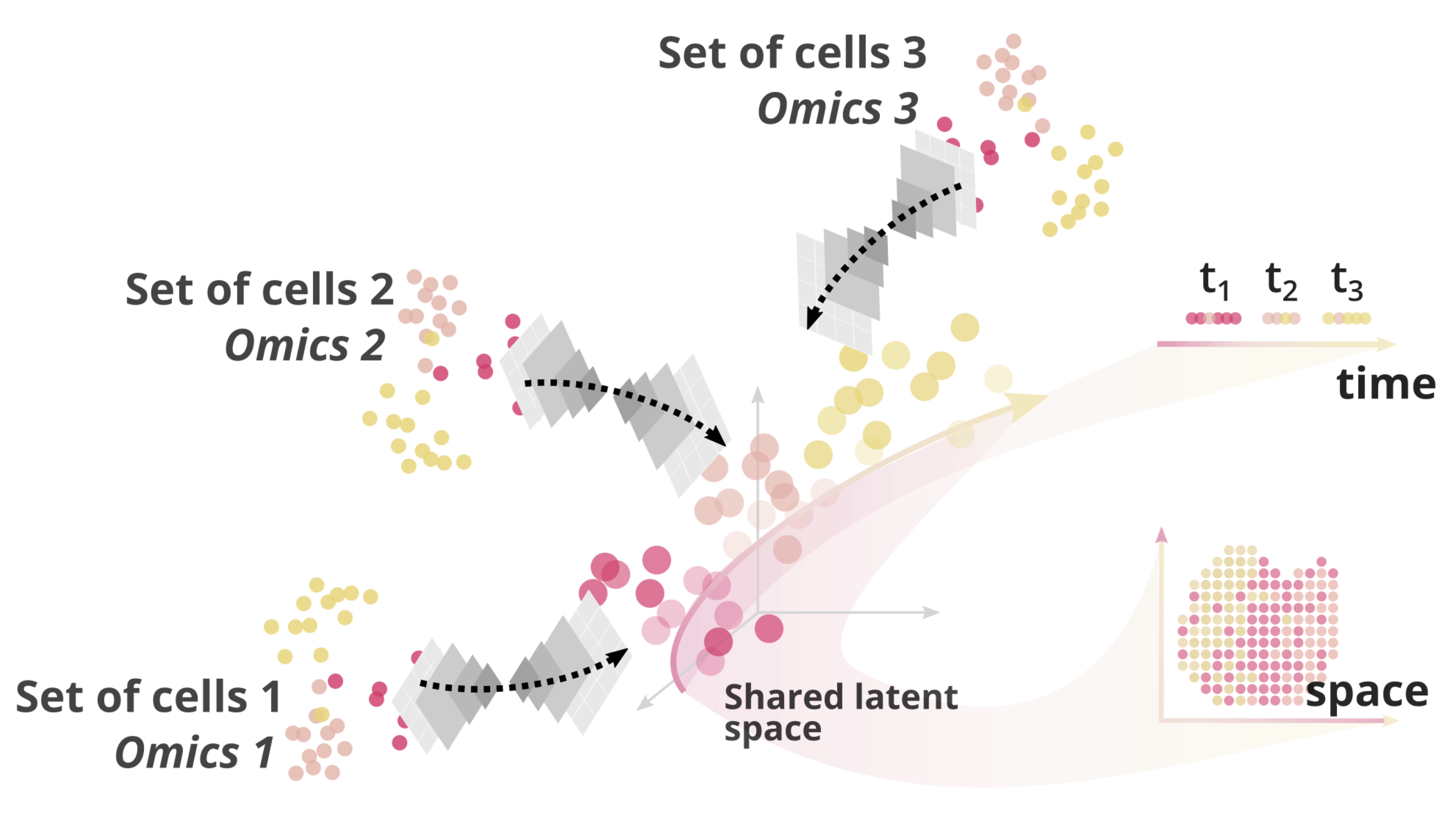
The introduction of high-throughput single-cell sequencing has produced a flood of data at the resolution of the single cell, including spatiotemporal information and different molecular facets of a cell, a.k.a. multi-omics. Their integration through MultiModal Learning (MML), aimed at combining multiple complementary views, offers great promise to understand the spatiotemporal phenotypic evolution of a cell and its molecular regulators. However, integrating multi-omics data across space and time is a huge computational challenge requiring radically new MML approaches.
The overall goal of MULTIview-CELL is thus to develop radically new MultiModal deep architectures enabling the pioneering integration of single-cell multi-omics data across space and time.
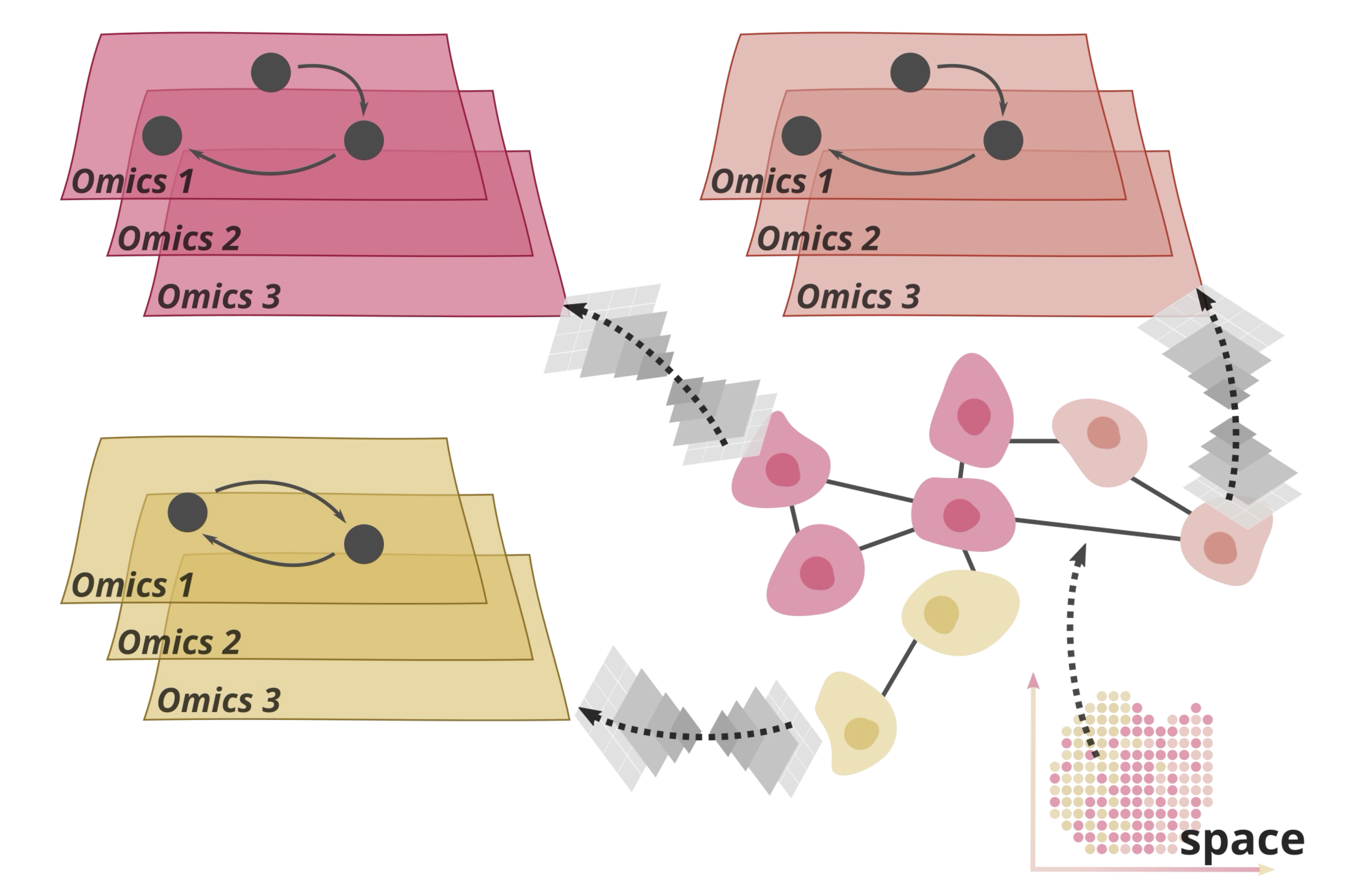
Aim 1. MULTIview-CELL will infer multimodal spatiotemporal phenotypic cell trajectories by combining back-translation, to allow the unsupervised dimensionality reduction of multimodal data, with a new Optimal Transport distance, allowing the spatiotemporal pairing of cells.
Aim2. MULTIview-CELL will then pinpoint the molecular regulators of such trajectories by combining new Graph Convolutional Networks with topological evolutions and Heterogeneous Multilayer Graphs, allowing the integration of graphs inferred from multimodal data (Aim2).
Aim3. Finally, all developed methods will be implemented in open-source software, with an emphasis on GPU-friendly scalable computations, a unique feature among existing single-cell tools.
These core contributions will impact Machine Learning, but more importantly, will have profound biological implications. The application of the tools developed to cutting-edge single-cell data from muscle stem cells will lead to new biological hypotheses on their heterogeneity and crosstalk, to be validated through wet-lab experiments (Transversal Tasks). In addition, by allowing to answer longstanding questions on the spatiotemporal phenotypic evolution of a cell, MULTIview-CELL will catalyze the generation of crucial
knowledge in fundamental biology and it will be key to preventing disease onset or therapy resistance, thus impacting health, society and economy.
Project results
Publications :
Public outreach :
scPRINT : le premier modèle d’IA français pour déchiffrer les réseaux génétiques | CNRS Biologie
Laura Cantini : ERC Starting Grant à l’interface entre apprentissage automatique et biologie
Institut Pasteur video about scPRINT
Job opportunities
Postdoc position in Machine Learning for Integrative Genomics