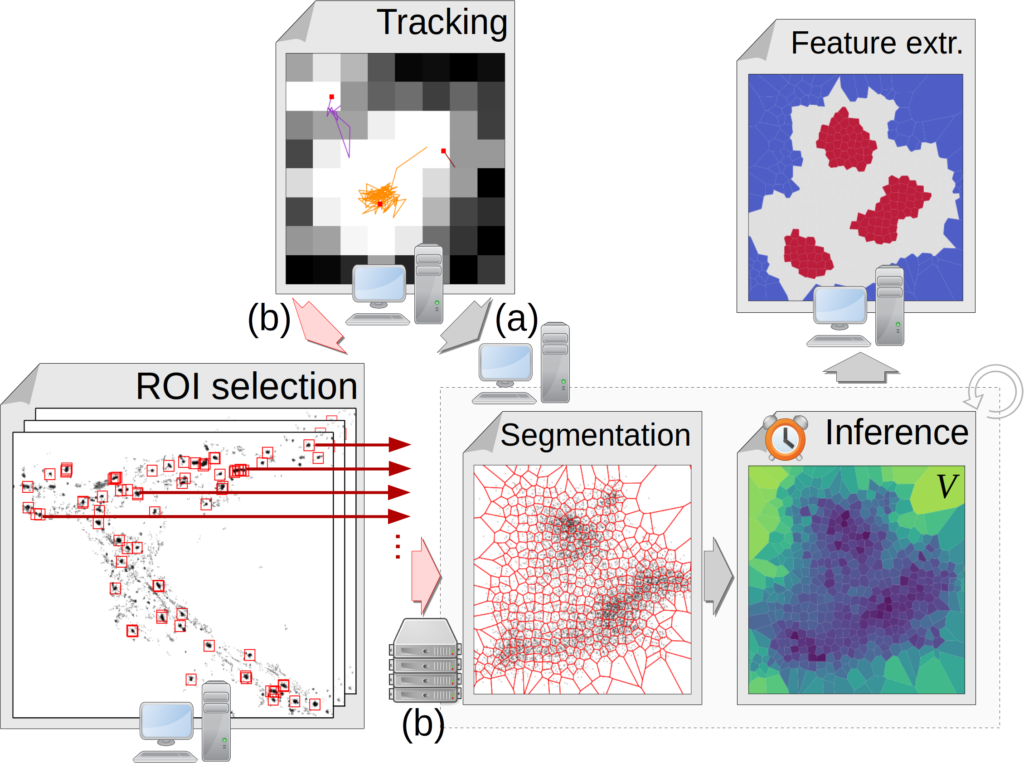
Présentation
Single-molecule localization microscopy (SMLM) allows studying the dynamics of molecules in cells and resolving the biophysical properties that underlie cellular function. With the continuously growing amount of data produced by every single experiment, the computational cost of quantifying these properties increases and becomes a major bottleneck in SMLM data analysis. Mining these data requires an integrated and efficient analysis toolbox.
TRamWAy is an open-source Python library that features:
- a conservative tracking procedure for localization data,
- a range of sampling techniques for meshing the spatio-temporal support of the data,
- computationally efficient solvers for inverse models, with the option of plugging in user-defined functions,
- a collection of analysis tools and a simple web-based interface.
It can additionally process in parallel the many data files in a dataset and regions of interest in each file. For example, from a local IPython notebook, it can connect to a remote HPC cluster, submit jobs and retrieve the generated data files onto the local host.