Présentation
Guy-Franck Richard, Alix Kerrest
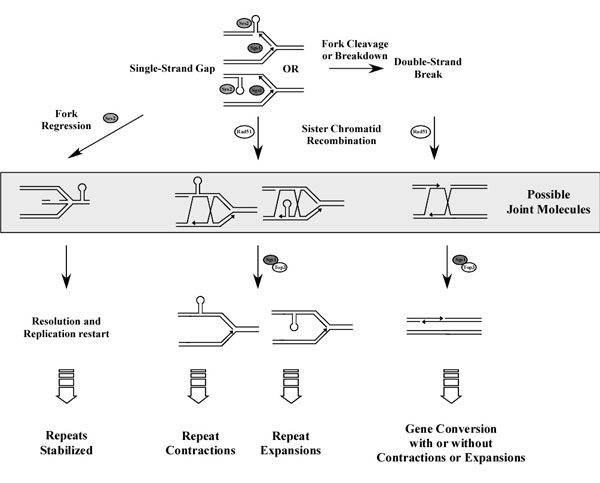
We are interested in understanding how DNA tandem repeats, like microsatellites and minisatellites are born, evolve and disappear. Trinucleotide repeats, a particular class of microsatellites, are involved in a growing number of human neurological disorders, including X-fragile, Huntington disease and several types of ataxias. The pathology is always associated to the large expansion of a trinucleotide repeat within or near a gene. We have shown that these expansions, in yeast, occur both during DNA replication and homologous recombination. We have isolated two genes, SGS1 and SRS2 (encoding DNA helicases conserved from bacteria to humans) that are involved in stabilizing trinucleotide repeats. We now try to understand the tight links between replication and homologous recombination and how these two metabolic pathways interact with each other.
Minisatellites are longer tandem repeats, also found in all eukaryotic genomes. By comparative genomics, we have shown that they evolve faster than the genes containing them. The mechanisms underlying birth, fast evolution and death of minisatellites are currently under investigation.
Our main questions are:
- How do DNA helicases interplay in stabilizing tandem repeats in vivo?
- How do replication and homologous recombination interact with each other during the cell cycle?
- What are the molecular mechanisms involved in the de novo formation of tandem repeats?
In order to adress these questions, we use classical reverse genetics and molecular biology approaches, such as conditional mutants, DNA two-dimensional gel electrophoreses and chromatin immunoprecipitation.