Présentation
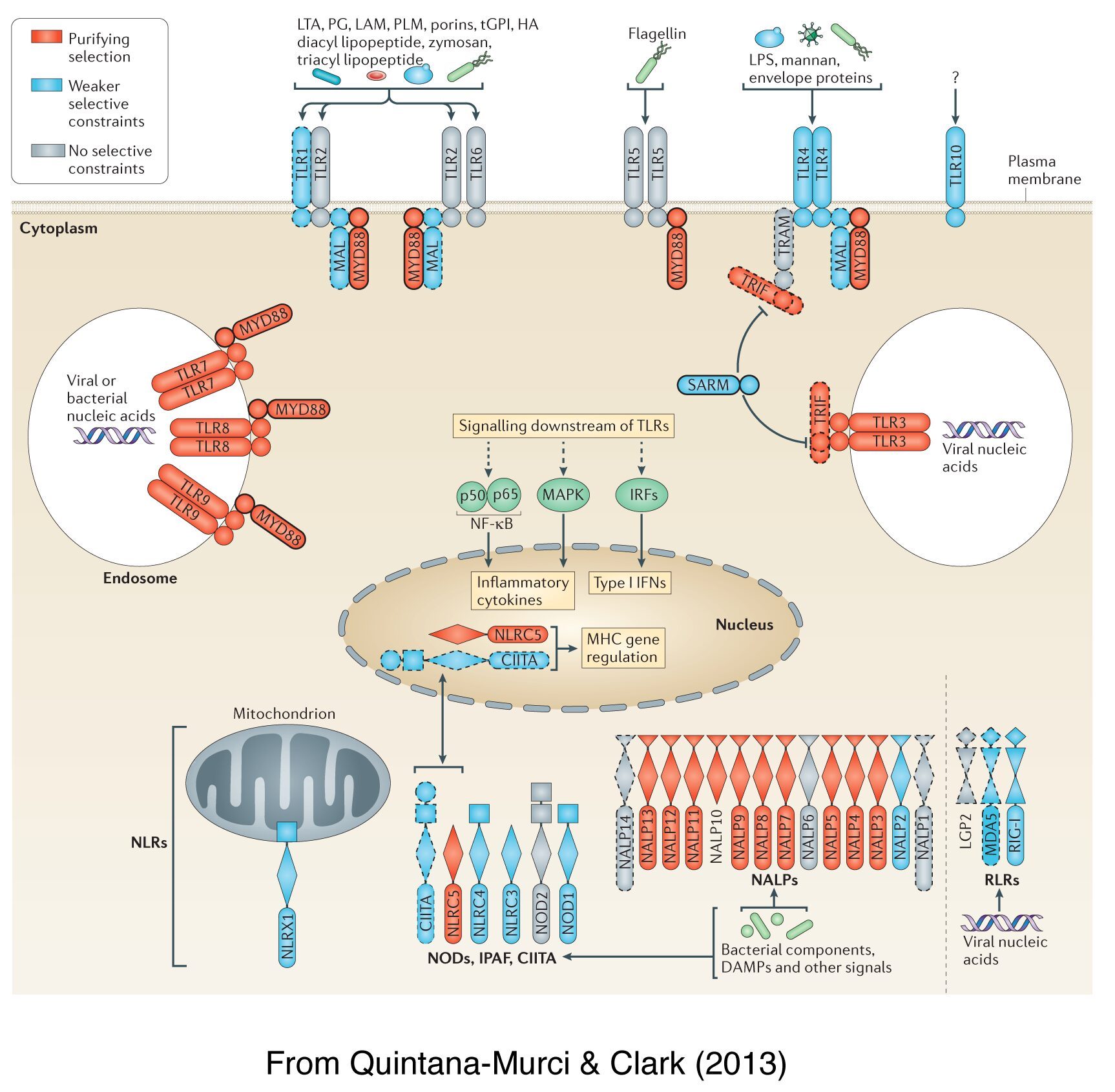
Inferences concerning the action of selection provide a powerful tool for predicting regions of the genome associated with disease. As infectious diseases exert strong selection pressures, the identification of immunity-related genes targeted by selection may provide insight into immunological defence mechanisms and highlight pathways playing an important role in pathogen resistance. Over the past years, we have focused on the dissection of how pathogen-driven selection has impacted the diversity of genes involved in innate immunity, including families of microbial sensors – such as the TLRs, NLRs and RLRs – as well as effector molecules – such as interferons. Our population genetic analyses have shown that innate immunity microbial sensors and other molecules involved in immunity to infection differ widely in their biological relevance, and highlighted evolutionarily important determinants of host immune responsiveness in the natural setting. Our work has also demonstrated that a G6PD-deficiency mutation is under strong positive selection in Southeast Asian populations, and provided a direct link between this mutation and protection against Plasmodium vivax rather than P. falciparum. This selection event coincides with the time at which rice started to be extensively cultured in the region — generating breeding grounds for mosquitoes — providing a link between environmental changes, natural selection and human health.