Présentation
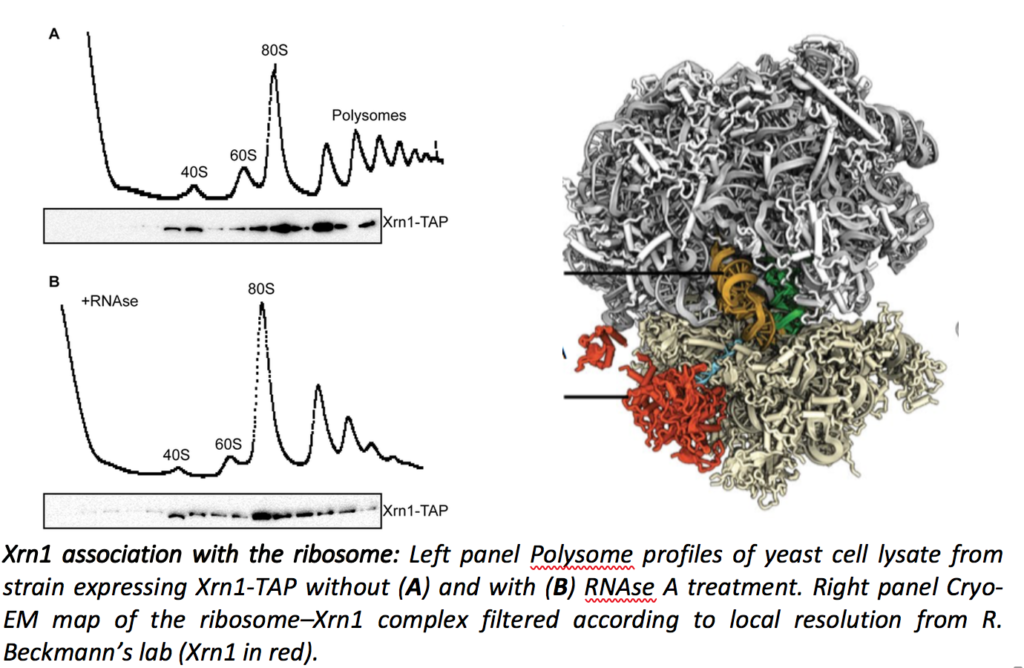
Cellular mRNAs are subjected to a balance between synthesis and degradation, which determines the shape of the transcriptome and greatly participates to protein homeostasis. In addition to the general degradation pathways, which ensure the turnover of each mRNA, a number of quality control mechanisms target aberrant RNAs for degradation. mRNA turnover has been shown to occur co-translationally, mainly by the Xrn1-dependent 5’ to 3’ degradation pathway. In collaboration with Roland Beckmann’s lab in Munich, we showed that Xrn1 is directly associated with the ribosome (Tesina et al., Nat. Struct. Mol. Biol. 2019).
Cytoplasmic mRNA degradation also involves, although to a lesser extent, the exosome/SKI-dependent 3’ to 5’ pathway. We recently showed that the SKI complex directly interacts with ribosomes and that this interaction takes place during the Non-Stop mRNA decay as well as in the general 3’ to 5’ mRNA degradation pathway (Schmidt et al., Science. 2018).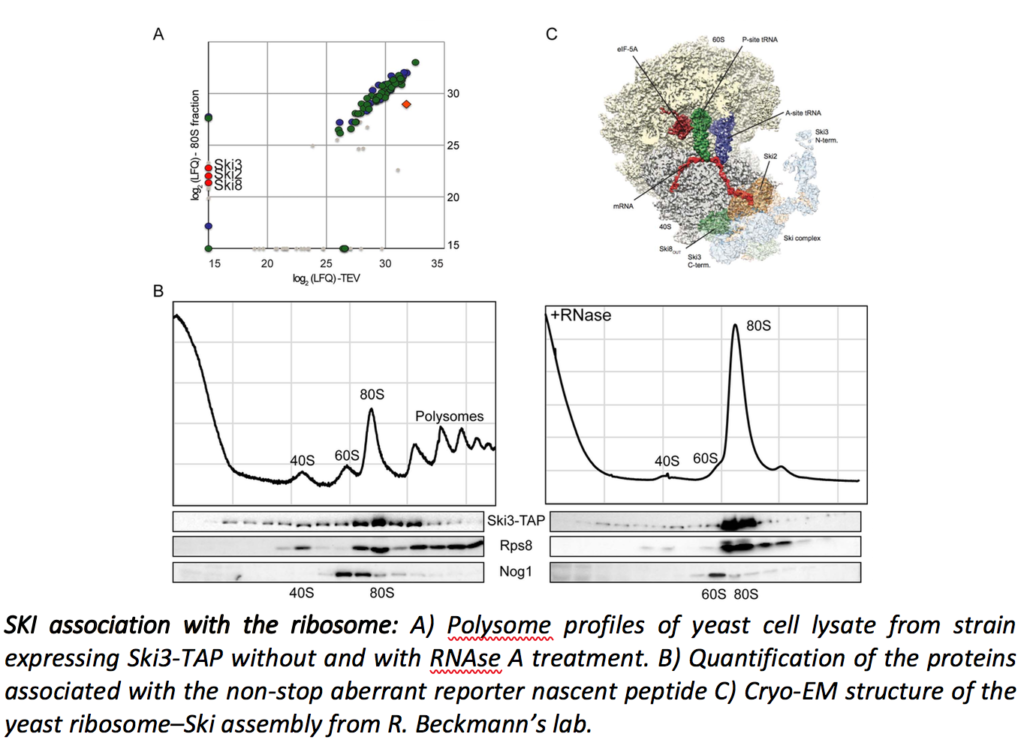
This seems paradoxical because the SKI complex was previously shown to be required for the efficient degradation of mRNA 3’UTRs, mainly devoid of ribosomes. Recently, we characterized, in Saccharomyces cerevisiae, a new complex involved in the degradation of the ribosomes-free RNA 3’UTRs and cytoplasmic lncRNAs such as XUTs and SUTs. We propose a model to describe the role of this complex (Zhang et al., EMBO J. 2019).
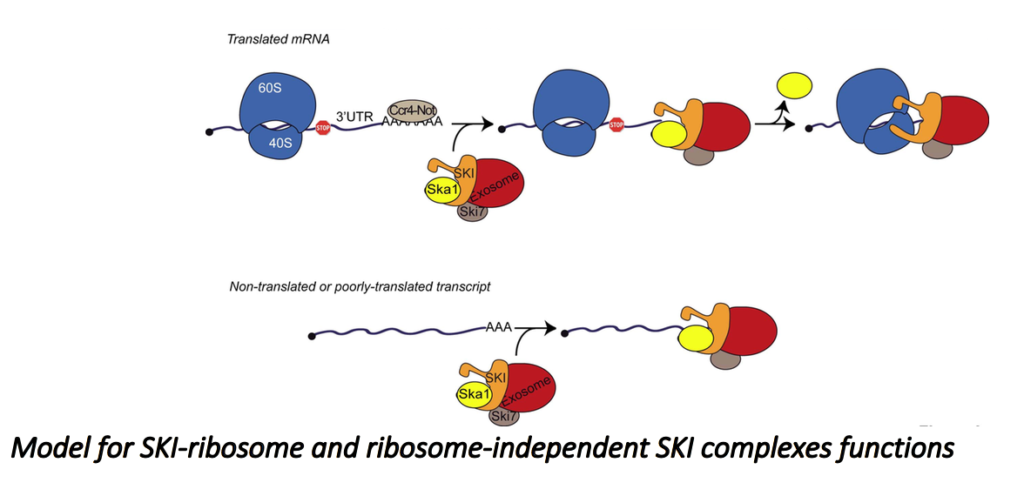 We are now better characterizing the SKI/Ska1 complex and exploring the two exonucleolytic pathways.
We are now better characterizing the SKI/Ska1 complex and exploring the two exonucleolytic pathways.