Présentation
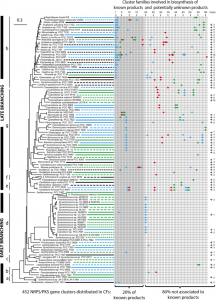
Cyanobacteria produce a large variety of bioactive compounds, some of which are toxic. But little is know about this natural product capacity at the phylum level. Our phylum-wide study highlighted the benefits of diversity-driven genome sequencing and foregrounded the huge diversity of gene clusters for secondary metabolite biosynthesis (Calteau et al., 2014). The investigation of these clusters present in various cyanobacteria gave deeper insights into the diversity and evolution of these ribosomally-dependent and non-ribosomally produced metabolites. Moreover the biosynthetic gene clusters can be organized very differently from one group of producing cyanobacteria to another (Dittmann et al., 2015)
Studying the clusters at the genetic level permitted to our colleagues in chemical and biochemical laboratories to more directly attempt the characterization of metabolites and variants produced by the PCC strains: Aeruginoguanidine/microguanidine/microguanidine amide (Pancrace et al., 2018); aranazoles (Moosmann et al., 2018); novel olefins (Zhu et al., 2018); merosterol (Moosmann et al., 2017); hassallidin E (Pancrace et al., 2017); tolytoxin and luminaolide B (Ueoka et al., 2015), variants of Microcystis natural compounds (Briand et al., 2016). With the first cultivated sources of actin-binding metabolites among the PCC strains (Ueoka et al., 2015), we now aim to study the potential use of these metabolites for medical applications.
With this multidisciplinary approach combining genomic, microbiology and chemistry done in the frame of collaborations, we aim to better understand the pathway diversification of the bioactive compounds and the evolutionary forces driving this remarkable genetic and chemical diversity. This way, we were able to reveal from Cyanobacteria a new family of architecturally and functionally distinct enzymes that exhibit high regioselectivity, substrate promiscuity, and irreversible action to introduce diverse D-amino acid patterns into peptide natural products (Morikana et al., 2014). We further examined these pathways and revealed a three-players synthesis for the various epimerisation of the final compounds (Morinaka et al., 2016). Finally, we reported a genome-guided discovery of bacterial pathways that posttranslationally create β-amino acid-containing products (Morinaka et al., 2018).
On going on this project:
- The 36-month project CYPHER funded by the ANR to better study the cyanobacterial toxin production and photoprotection processes in a changing environment (collaboration with Pr C. Bernard, MNHN & Dr D. Kirilovski, CNRS/CEA).