Présentation
Chikungunya virus p62-E1 envelope glycoproteins
The Chikungunya virus (CHIKV) is an emerging mosquito-borne alphavirus that has caused widespread outbreaks of a debilitating human disease in the last 5 years. CHIKV invasion of susceptible cells is mediated by two viral glycoproteins, E1 and E2, which carry the main antigenic determinants and form an icosahedral shell at the virion surface. The glycoprotein E2, derived from furin cleavage of the p62 precursor into E3 and E2, is responsible for receptor binding, and E1 for membrane fusion. In the context of a concerted multidisciplinary effort to understand the biology of CHIKV, we report here the crystal structures of the precursor p62-E1 heterodimer and of the mature E3-E2-E1 glycoprotein complex.
The resulting atomic models allow the synthesis of a wealth of genetic, biochemical, immunological and electron microscopy data accumulated over the years on alphaviruses in general. This combination yields a detailed picture of the functional architecture of the 25 MegaDalton alphavirus surface glycoprotein shell. Together with the report on the structure of the Sindbis virus E2/E1 heterodimer at acidic pH3 (Li et al, Nature, 2010), our structure provides new insight into the acid-triggered conformational change on the virus particle and its built-in inhibition mechanism in the immature complex.
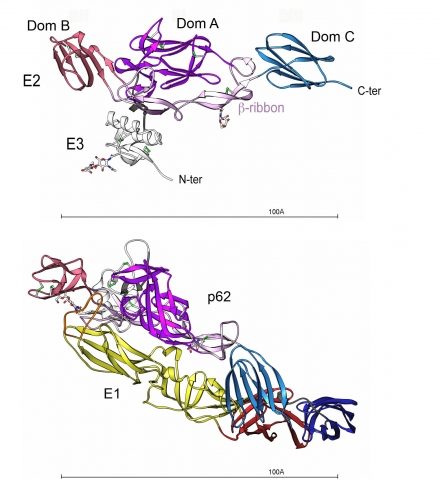
Structure of the p62-E1 heterodimer.
Top view. p62 molecule : E3 is colored white and E2 domain A purple, B red, C blue and the beta-ribbon pink. The N-linked glycans are shown in ball and stick, colored according to atom type. The disulfides are depicted as green sticks.
Bottom view (90° from the top view). p62-E1 heterodimer : E1 domains I, II and III are shown in dark red, yellow and dark blue, respectively, and the fusion loop is colored in orange.
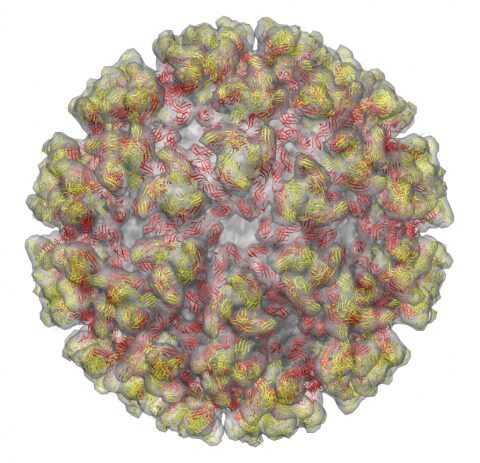
The alphavirus T=4 icosahedral surface glycoprotein shell.
The atomic model of the 240 chikungunya E2-E1 heterodimers are arranged as 80 spikes.
E2 is colored in yellow and E1 is in red.
The E2-E1 complex is fitted into the cryo-EM density of sindbis virion
(cryo-EM from: Mukhopadhyay S et al., Structure 2006).
Glycoprotein organization of chikungunya virus particles revealed by X-ray crystallography.
Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA.
Nature 2010 Dec 2;468:709-712.
This work was funded by the French ‘Agence Nationale de la Recherche’ (ANR, DENtry, program ‘Microbiologie, Infections et Immunité’), by Merck-Serono, by the Pediatric Dengue Vaccine Initiative (PDVI), by the Institut Pasteur ‘Programme Transversal de Recherche’ (CHIKV, PTR n°201) and by recurrent funding from Institut Pasteur and CNRS. V.J.E. was supported by a Marie Curie fellowship through the European Union Research Training Network program ‘‘Intrapath’’.
Press release (in english) (in french)
News and Views: ’Structural biology: An alphavirus puzzle solved’ by Margaret Kielian.
Nature. 2010 Dec 2;468:645-646.



