Présentation
Nucleoside analogues as potential antitumor agents.
This project aims to characterize and validate the nucleotide N-hydrolase DNPH1 as a new target in oncology. Inhibition of Rcl alone or in combination with other chemotherapeutic drugs should lead to the development of a new strategy to inhibit cancer proliferation, as well as new diagnostic tools to predict the potential malignancy of cancers. This program was supported by the Fondation pour la Recherche Médicale (Chimie pour la Médecine, DCM20111223063).
Deregulated expression of c-myc plays a significant role in human cancer development. Among the putative c-Myc-responsive genes, rcl (C6orf108) has been recognized as a growth-related gene. rcl is among the top 50 genes that allow to distinguish benign and malignant prostate tissues. The up-regulation of rcl has been observed in various human cancers, including breast and prostate, colon carcinoma, as well as chronic lymphocytic leukemia. In addition, rcl expression increases significantly with the tumor grade, strongly connecting rcl to breast tumorigenesis, but direct evidence for the role of Rcl in cancer development is still lacking.
The rcl encoded protein, recently renamed DNPH1, was identified as an original nucleotide N-hydrolase that catalyzes the glycosidic cleavage of a 2′-deoxynucleoside 5′-monophosphate into a free base and 2-deoxyribose-5-phosphate, while the corresponding ribonucleotide acts as a competitive inhibitor (Ghiorghi, K. et al., 2007). Its role in nucleotide catabolism has suggested that it might fuel cancer growth. The NMR structures of rat DNPH1, in the apo form and in complex with GMP, provided the first structural insights into the substrate recognition (Yang et al., 2009). The substrate specificity was analyzed using rational site-directed mutagenesis and a small set of nucleotide analogues (Dupouy et al., 2010).
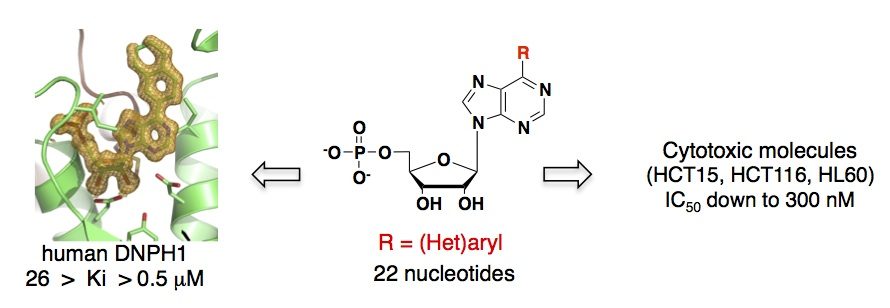
A systematic chemical exploration of critical binding determinants through the synthesis of purine nucleotide analogues allowed us to identify potent inhibitors of DNPH1 and gain better insight into protein-ligand interactions. First, a series of N6-substituted AMPs, including some cytotoxic cytokinin ribosides as 5’-monophosphate derivatives, was synthesized allowing the kinetic, thermodynamic and structural characterizations of the rat enzyme. These N6-substituted AMP derivatives were found to inhibit rat DNPH1 in the micromolar range, supporting a possible implication of DNPH1 in their activity (Amiable et al. PLOS One, 2013). The crystallographic data of rat DNPH1 in complex with three N6-substituted AMP derivatives revealed tight interactions with the ribose moiety and phosphate group, while the modified purines showed little interactions with the protein (Amiable et al., 2013, Padilla et al., 2013).
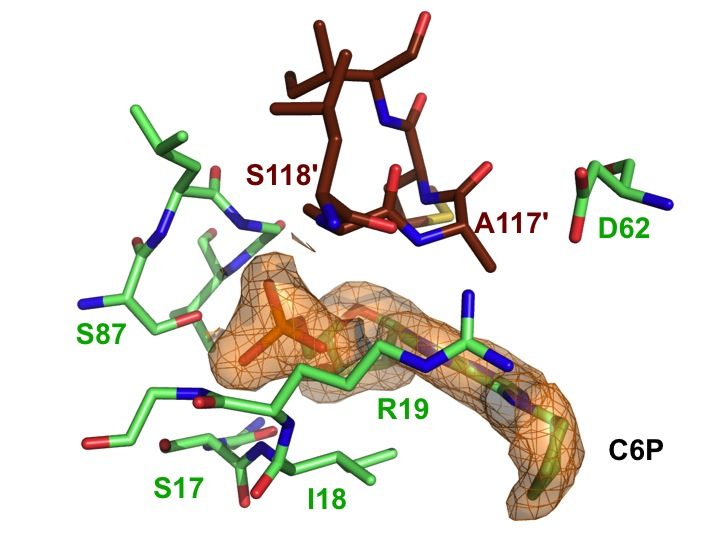
The SAR study was further pursued by introducing less flexible substituents (as arenes and heteroarenes) at the 6 position of the purine core. A diverse set of 6-aryl- and 6-heteroarylpurine ribosides as 5′-monophosphate derivatives was synthesized in good overall yields, via Suzuki-Miyaura cross-coupling reactions. Biochemical studies revealed competitive inhibitors of both rat and human recombinant DNPH1 enzymes, with Ki values in the low micromolar range. The best inhibitors exhibited in vitro cytotoxicity against human colon cancer (HCT15, HCT116) and human promyelocytic leukemia (HL60) cell lines, with IC50 values in the low micromolar range (Amiable et al., 2014). Cytotoxicity relies on their phosphorylation by adenosine kinase, and probably their accumulation as monophosphorylated form, as shown previously for active cytokinins and triciribine. Finally, one nucleotide from this series cocrystallized with human DNPH1, providing the first detailled 3D structure of the human target (Amiable et al., 2014). These data are now used for the development of more potent inhibitors.