Présentation
Targeting antimicrobial resistance through bacterial adhesin research
Strains, Diagnostics, and Variants
StraDiVarious is a European doctoral network studying strain variants of ESKAPE pathogens to develop rapid diagnostics and identify therapeutic targets.
Our research hypothesis is: small structural variations in virulence factors modulate the virulence of different bacterial strains of the same species, and, in turn, the course of infection and disease outcome. Despite the high medical need, this variation within pathogenic species is still underexplored. Diagnostic workflows rarely distinguish between such strains and variants. Antimicrobial therapy thus currently often makes no difference between highly virulent (killer) and low-virulent (commensal or colonizer) strains. Rapid distinction of these strains would be a game-changer wherever antibiotics are used empirically (e.g. intensive care, treating the elderly), allowing for much more targeted treatment. Untargeted antimicrobial treatment also leads to the ‘blooming’ of antibiotic-resistant but clinically silent bacteria in the gut. Antibiotic stewardship based on the pathogenic potential would thus reduce unnecessary antibiotic use and the risk of antimicrobial resistance (AMR).
Our work is interdisciplinary and collaborative – microbiologists, clinicians, bioinformaticians, structural biologists, cell biologists and bioengineers work on different aspects of bacterial strains and variants. Or current focus are bacterial adhesins, the virulence factors that are essential for first contact with the infected host, and for establishing and maintaining an infection by attaching to different cells and surfaces.
15 doctoral candidates will be hired and will do their PhD during 2026-2029.
The call for applicants to the 15 Doctoral Candidates in StraDiVarious opens November 12 2025
https://www.stradivarious.eu/apply
My group is proposing one project:
DC7: Impact of microenvironment on colonisation and virulence of enteric bacteria
WP2: Adhesin-mediated host cell interaction in infections
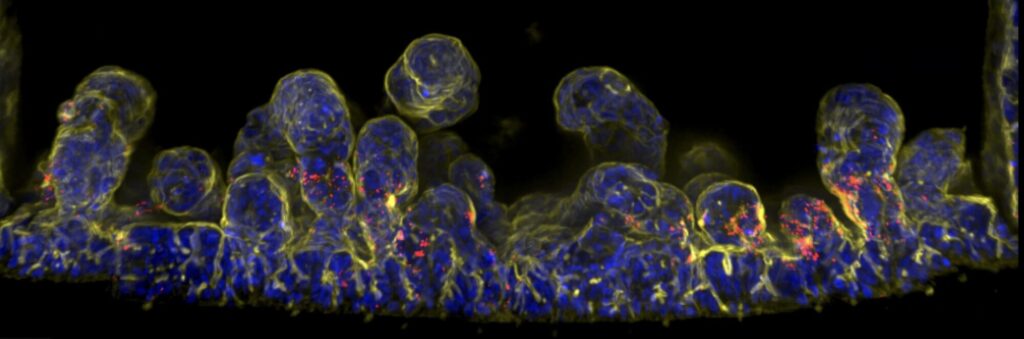
The PhD student will work on the impact of the microenvironment of the human gut on colonisation and virulence of enteric bacterial pathogens (EHEC). In this project you will use a highly physiological human colon model, combining patient-derived colonic organoids with organ-on-chip (OoC) and enabling to modulate the key gut environmental cues: mechanical stimulation (peristalsis and shear stress) and microbiota metabolites. Using several EHEC strains based on their differences in adhesins or Type IV pili, you will help to characterize the colonization, adhesion of these variants, and the consequence of the gut environmental cues on the EHEC virulence.
The work is in close collaboration with other groups in the network interested in host cell receptors, and in anti-adhesion drug development. The position thus offers the opportunity of internships with both academic and industrial partners.
Methods will include the generation of human colonic organoids, barrier maturation in OoC, infection assays in OoC modulating mechanical forces and microbiota metabolites, live and fixed imaging quantifying the bacterial adhesion and colonization, CFU counting, barrier leakage assays, immunofluorescence and RTqPCR analysing the downstream impact of infections on the colonic barriers.
Key Dates
- 12 Nov 2025 applications open
- 6 Jan 2026 application deadline
- 9, 13 Feb 2026 online interviews take place; candidates need to be available on both days.
- By March 2026 offers sent to successful candidates
- Aug – Sep 2026 Commencement of posts