About
The past years have been marked by a leap forward in our understanding of the genetic basis of many diseases. In particular, genome-wide association studies (GWAS) have provided insight into the “genetic architecture” of immune-mediated inflammatory diseases. These studies have highlighted many genes linked to signaling pathways that were not known to be involved in pathogenesis, and have pointed to new directions in the study of disease mechanisms. GWAS also provided conclusive evidence for a key role of the immune system in the pathogenesis of these diseases, because many of the identified loci map to genes that regulate a large variety of immune processes.
The current challenge is to correlate these genetic variants with the effector mechanisms implicated in pathogenesis, to allow translation of the genetic data into relevant diagnostics and innovative treatment strategies.
We have investigated the impact of genetic polymorphisms in genes of the IL-23 signaling pathway that have been linked to spondyloarthritis (SpA) on the effector functions of CD4+ T cells from (SpA) patients. We measured the expression levels of Th17 and Th1 cytokines and transcription factors in CD4+ T cells isolated from SpA patients and correlated them with the patients’ genotype at loci genetically associated with spondyloarthritis. We found that SpA patients carrying risk-associated alleles of genes in the IL-23/IL-17 pathway (such as IL23R, IL12B and CCR6) expressed the highest levels of genes involved in the differentiation and function of Th1 and Th17 cells, whereas the presence of protective alleles was associated with low-level expression of these genes (Fig. 1A and B). In contrast, variation at loci genetically linked to SpA, but not associated with the IL-23 pathway (such as ERAP1 and ANTXR2), did not correlate with expression of Th1 and Th17 genes, suggesting that these SNPs may contribute to SpA pathogenesis through distinct cellular mechanisms (Fig. 1C).
These data show that genetic variation at multiple loci associated within the IL-23 signaling pathway affects CD4+ effector functions in SpA patients.
To determine a potential combinatorial effect of SNPs at genes associated with the IL-23 signaling pathway on Th1 and Th17 cell functions, we correlated the expression levels of inflammatory cytokines with the genetic risk associated with 5 alleles targeting the IL-23 pathway using a multiplicative model. We found that patients having a higher cumulative risk express significantly higher levels of Th1 and Th17 marker genes than patients with a lower cumulative risk. To determine whether this is restricted to SNPs at loci associated with the IL-23 signaling pathway, we performed the same analysis using 5 SNPs associated with SpA, but not targeting molecules in the IL-23 pathway. This analysis revealed no differences in the two groups, suggesting that genetic risk associated with SNPs in the IL-23 pathway, but not in other signaling pathways, correlates with Th1 and Th17 effector functions.
These data show that the combinatorial action of multiple SNPs at distinct loci, rather than a single genetic variant, determines the immune phenotype of SpA patients. Our results demonstrate, for the first time, a link between disease-associated genetic variants and defined functions of immune cell populations involved in the pathogenic mechanisms of inflammatory disease. These findings establish a novel conceptual and experimental framework to delineate the mechanisms by which genetic variants contribute to pathology (Coffre et al, 2013).
This project is performed in collaboration the team of Pr. M. Dougados, Director of the National Reference Center for SpA in France and of the Service de Rhumatologie B at Hôpital Cochin and with the Center for Human Immunology (CIH) in the immunology department.
Supplementary data for this article can be downloaded here
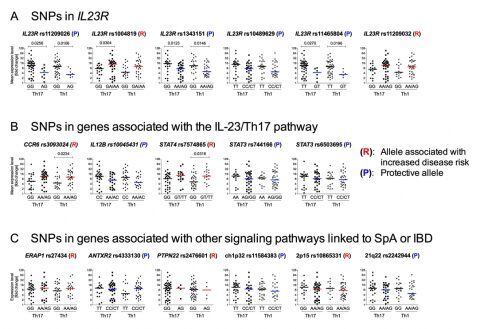
Figure 1. Overview of the effect of genetic variation at loci linked to SpA on Th17 and Th1 cell functions.
SNPs linked to SpA at IL23R (A), genes associated with the IL-23/Th17 axis such as CCR6, IL12B, STAT3, and STAT4 (B), and at genes linked to SpA but not associated with the IL-23 signaling pathway (C) were selected from the literature. We also included SNP rs7574865 at STAT4 and rs2476601 at PTPN22 that are strongly associated with increased disease risk in RA and SLE. Major and minor alleles are indicated below the X-axis. (P) and (R) indicate protective and risk-associated minor alleles, respectively. To provide a better overview, the expression of the 4 Th17 marker genes (IL17A, IL17F, IL23R and RORC) and 4 Th1 markers (IFNG, TNFA, IL12RB2 and TBX21) were combined after normalization and represented as “mean expression level” and plotted on the Y-axis (indicated by Th17 and Th1 below the X-axis). Mean expression levels were calculated by setting the highest fold-change value for each gene to 100 and scaling the other values accordingly, followed by calculating the mean value for Th17 marker genes and Th1 markers (indicated by “Th17” and “Th1” below the X-axis). (Figure S4 in Coffre et al, 2013).