About
Non-sense mediated mRNA decay (NMD) is a fundamental and conserved mechanism with a major role in shaping the levels of individual RNAs in a eukaryotic cell by efficiently removing RNAs in which a stop codon occurs prematurely during translation. While three proteins, Upf1, Upf2 and Upf3 constitute the core of the machinery involved in this process, the process hijacks normal degradation pathways for mRNAs, including the decapping and deadenylation machineries. Understanding NMD mechanisms thus can help understanding fundamental processes involved in mRNA stability in eukaryotes.
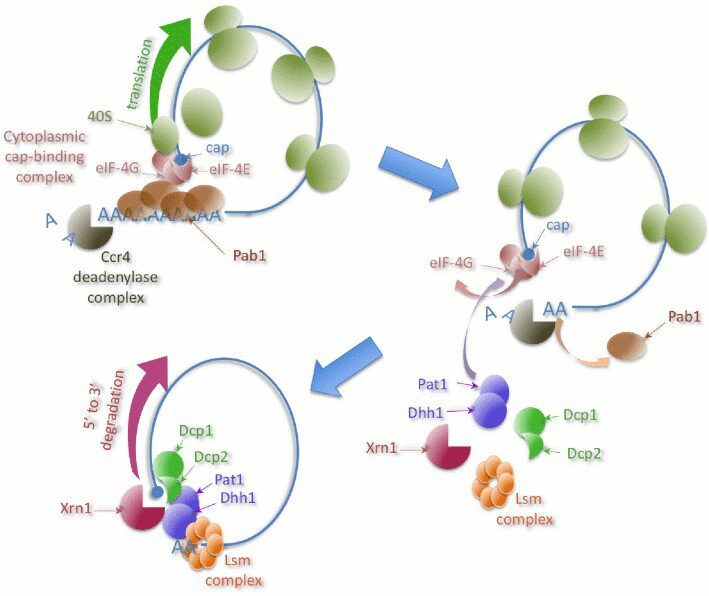
One of the major steps in mRNA degradation is the decapping reaction, usually triggered by a reduction in the size of the poly(A) tail. Sometimes, decapping can occur without deadenylation. We identified a mechanism of deadenylation-independent mRNA decapping and degradation that involves the protein Edc3 – a factor physically and genetically linked to the decapping machinery. The auto-regulation mechanism involved in the fine-tuning of the RPS28B mRNA levels depends on Edc3 (Badis et al., Mol Cell 2004). We analyzed other candidate proteins that are involved either in general decapping or in the regulation of specific transcripts levels. The general method for quantitative tests of the effect of combining mutations on a large scale, allow the identification of novel functional links in RNA synthesis and decay (Decourty et al., PNAS 2008).
We are currently interested in nonsense mediated mRNA decay (NMD) substrates and mechanisms. To better understand what are the main features of mRNA that are degraded through NMD, we tested a set of 600 reporters in a molecular barcode based assay. Surprisingly, while a long 3’UTR is a required and well known factor that favors NMD, we found that the length of the translated ORF also plays a role, with long ORF transcripts showing higher stability and less sensitivity to NMD Cell Rep 2014.
The approaches we use involve quantitative characterization of protein complexes and their associated RNA, to better understand the biochemistry of the NMD process, including the order of arrival and departure of various factors and the events that trigger major steps in the decay of mRNA through this pathway. The information obtained from these experiments is combined with data mining approaches in large scale results obtained by synthetic lethal screens and other published resources.
For the most recent results on NMD complexes, please refer to our recent pre-print manuscript (Dehecq et al., 2018). or the corresponding publication in the EMBO Journal. To explore the quantitative data: http://hub05.hosting.pasteur.fr/NMD_complexes/
ANR-14-CE10-0014, CLEANMD 2014-2018, DEFineNMD 2019-2023