Contact : mathis.fleury@pasteur.fr
Beginning Feb-Mar 2025
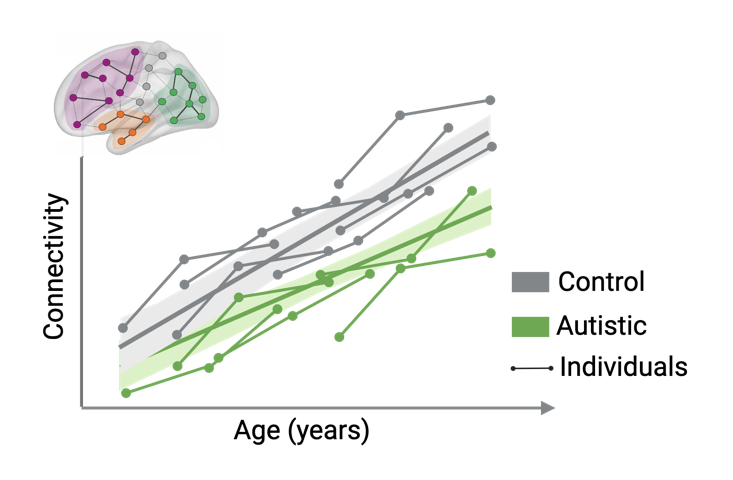
From childhood to adult, brain connectivity development in autistic individuals.
The genetic architecture of autism is complex, ranging from monogenic forms involving a single rare deleterious variant, e.g. a single nucleotide variant (SNV) or copy-number variant (CNV) of the condition to highly polygenic forms resulting from the additive effect of thousands of small effect single nucleotide polymorphisms (SNP). Our current knowledge of the biological pathways associated with autism and intermediate brain phenotype (e.g. macro/microcephaly) is mostly based on the identification of rare variants impacting proteincoding genes. There are more than 200 genes associated with autism, and recent studies reported the combined effect of rare and common variants distributed throughout the genome1. Moreover, phenotypic outcome could be the consequence of a collective contribution of regulatory and protein-coding variants2,3.
Neuroimaging genomic studies of autism have mainly adopted ‘top-down’ approaches, starting with the behavioural diagnosis, and moving down to intermediate brain phenotypes and underlying genetic factors. Such studies have identified patterns of neuroimaging alterations with relatively small effect-sizes and an extreme polygenic architecture4. In stark contrast, bottom-up studies are based on the presence of genetic susceptibility factors for autism with relatively higher impact, independently from the behavioural diagnosis5.
Advanced EEG techniques (e.g., connectivity, complexity and entropy features) are suited to investigate macroscopic neuronal circuit function and maturation. EEG and other electrophysiological techniques are noninvasive and direct measures of neuronal activity with a high temporal resolution enabling the evaluation of brain dynamics and brain rhythms. These techniques show promise for identifying biomarkers of atypical brain development6. Differentiating normative individual variability in brain development from pathological changes induced by autism is essential to identify children who deviate from typical brain development and are thus at higher risk for long-term deficits. Moreover, understanding resilience—the ability of some individuals to maintain typical brain development despite genetic susceptibility—is crucial. Achieving this requires integrating advanced EEG with longitudinal statistical methods sensitive to individual changes.
Aims and hypotheses:
Examine the long-term impact of autism on the development of brain networks. Longitudinal statistics and advanced EEG techniques will be combined to model the developmental trajectories of brain networks during adolescence, simultaneously evaluating intra- and interindividual variability as well as resilience to genetic and environmental variations.
Internship Opportunity:
This internship focuses on analyzing autism spectrum disorders (ASD) using EEG connectivity data and incorporating genetic analysis. It will be conducted within the GHFC team (Human Genetics and Cognitive Functions) at Institut Pasteur, led by Pr. Thomas Bourgeron.
The intern will be working with data from LEAP/InovAND dataset, an extensive longitudinal database that includes 600 adolescents without diagnosis of autism and 250 individuals with autism.
The internship comprises the following key technical responsibilities:
1. Data Preprocessing: The intern will preprocess the longitudinal EEG data, involving steps such as noise reduction, motion correction, and normalization, to prepare the data for analysis.
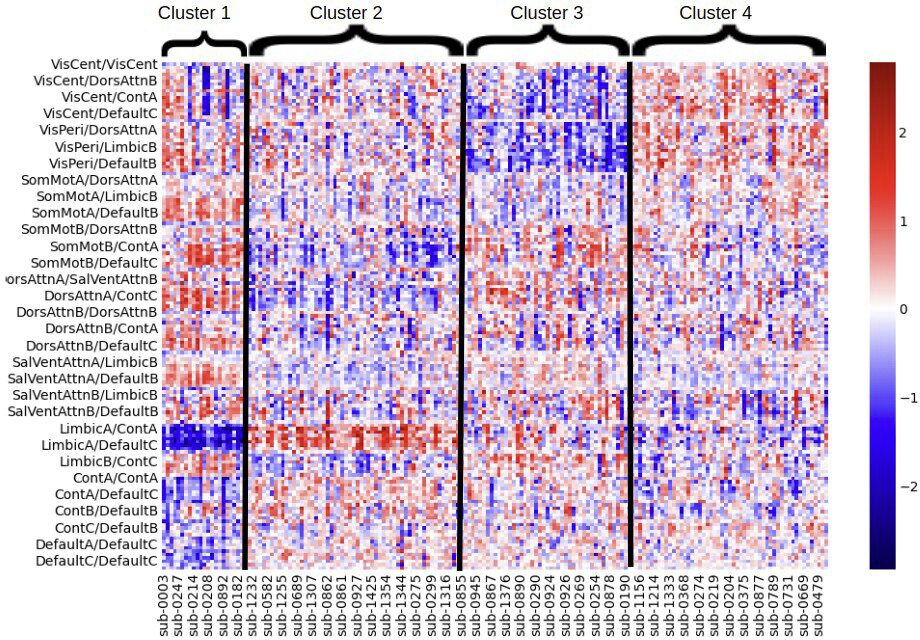
2. Machine Learning and Clustering Analysis: Using machine learning algorithms (Scikitlearn library), the intern will perform clustering to differentiate between ASD and typically developing individuals based on neural connectivity patterns6.
3. Genetic Data Integration: The final step involves integrating genetic information with the neuroimaging data. This will include analyzing genetic variants and their potential correlations with observed connectivity patterns in ASD, contributing to the emerging research in this area7.
It is designed for individuals with a background in neuroscience, data analysis, or related fields, who are interested in advanced neuroimaging techniques and genetic correlations in neurodevelopmental disorders. Candidates are expected to have a foundational understanding of neuroimaging techniques and data analysis (even though it is not mandatory), and an interest in the genetic aspects of neurodevelopmental disorders. The internship provides an opportunity to gain hands-on experience in EEG data preprocessing, machine learning-based clustering, and the integration of genetic and neuroimaging data. Additionally, the intern will gain experience with SLURM for managing and executing computations on a cluster.
References:
1. Rolland, T. et al. Phenotypic effects of genetic variants associated with autism. Nat Med (2023) doi:10.1038/s41591-023-02408-2.
2. Zhou, J. et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat Genet 51, 973–980 (2019).
3. Castel, S. E. et al. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat Genet 50, 1327–1334 (2018).
4. Lombardo, M. V., Lai, M. C. & Baron-Cohen, S. Big data approaches to decomposing heterogeneity across the autism spectrum. Molecular Psychiatry 2019 24:10 24, 1435–1450 (2019).
5. Moreau, C. A. et al. Mutations associated with neuropsychiatric conditions delineate functional brain connectivity dimensions contributing to autism and schizophrenia. Nature Communications 2020 11:1 11, 1–12 (2020).
6. Ren, P. et al. Stratifying ASD and characterizing the functional connectivity of subtypes in resting-state fMRI. Behavioural Brain Research 449, 114458 (2023).
7. Berto, S. et al. Association between resting-state functional brain connectivity and gene expression is altered in autism spectrum disorder. Nature Communications 2022 13:1 13, 1–11 (2022)