Context & Objectives
Genome-wide association studies (GWAS) – i.e systematic study of the link between millions of genetic variants and a trait in a sample of individuals- have been central to decipher the genetic factors of human traits and diseases. As of today, GWAS have reported over 150K associations for thousands of heritable traits (e.g. height, type 2 diabetes or cardiovascular diseases) 1 . This incredible amount of data has been used to unravel biological mechanisms implicated in human diseases, and chronic diseases in particular 2 . However, the shared genetic structure across human phenotypes and the associated functional component remain poorly understood. The development of innovative methods to address these questions is critical and extremely challenging. Our group has developed an internationally recognized expertise in multi-traits analysis methods –i.e. joint statistical tests of genetic variants on several complex traits- and their associated computational tools 3 . In particular, our recent work focused on how multi-traits analysis can be used to uncover functional pathways.
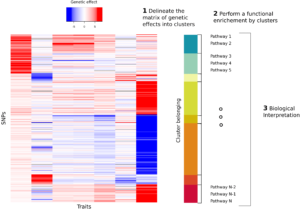
A primary target for our ongoing multi-trait genetic analyses is the genetic of metabolism and its link to diseases. Indeed, cholesterol and glucose plasma concentration are causally implicated in three of the deadliest chronic diseases globally 4 (Coronary heart disease, Strokes and diabetes). Hence, the utmost importance of better understanding the genetic of metabolism to improve and personalized treatment. The internship objective is to analyze a dataset of genetic effects on 159 metabolites (cholesterol, glucose…). The trainee will be asked 1) to find a synthetic decomposition of the genetic effect matrices 2) to map this decomposition to metabolic pathways (see figure below) and 3) to assess how this pathway annotation could be used to refine the computation of a Polygenic Risk Score. The trainee will review literature to apply state of the art method both for the matrix clustering and for the functional enrichment.
During his/her internship, the trainee will benefit from the stimulating scientific environment of the Pasteur institute including regular computational biology seminar and large group of bioinformatician and biostatistician
to exchange with.
Qualifications
The position requires advance knowledge in statistics and computer sciences. The applicants should therefore have substantial educational background in statistics/biostatistics, computer Science or other relevant disciplines. Prior knowledge in biology would be an asset. The call specifically addresses master 2 students and
3rd year engineer school students.
Additional informations
Interested applicants should send their curriculum vitae, a grade transcript, a brief cover letter, and contact information from at least one referee (e.g. teacher or previous internship mentor) to Dr. Hanna Julienne (hanna.julienne@pasteur.fr). More information about our group and the C3BI can be found here https://research.pasteur.fr/en/team/statistical-genetics/ and here https://research.pasteur.fr/en/center/c3bi/.
[gview file=”https://research.pasteur.fr/wp-content/uploads/2016/09/research_pasteur-statistical-genetics-sujet-stages-clustering.pdf”]



