About
Respiratory Syncytial Virus Nucleoprotein-RNA complex
Human respiratory syncytial virus (RSV) is the most important viral agent of pediatric respiratory tract disease worldwide, causing pneumonia and bronchiolitis in infants. No vaccine is currently available, and effective treatments have yet to be developed. RSV is a non-segmented, negative-strand RNA virus of the Paramyxoviridae family in the Mononegavirales order. The 15.2 kB genomic RNA contains 10 genes, four of which code for intracellular proteins involved in genome transcription, replication and particle budding that have orthologs in all Mononegavirales, namely N (nucleoprotein), P (phosphoprotein), M (matrix protein) and L (“Large” protein, the RNA polymerase). The RSV genomic RNA forms a nuclease-resistant helical ribonucleoprotein (RNP) complex with the N protein, termed nucleocapsid, which is used as template for RNA synthesis (transcription and replication) by the viral polymerase complex.
We solved the crystal structure of a decameric ring of the ribonucleoprotein complex of the RSV nucleocapsid (N) protein bound to RNA. The N-RNA ring features the RNA chain running in a basic surface groove, making a belt around the complex, displaying alternating rows of 4 and 3 stacked bases exposed to solvent and buried within the groove, respectively. The N subunit is organized as a core region containing two domains, N- and C-terminal (NTD and CTD), connected through a hinge region. The RNA groove is formed at the NTD/CTD interface, the inter-domain connection lines its internal side. This NTD-CTD core of the molecule has N- and C-terminal extensions, “N-arm” and “C-arm”, which interact laterally in the ring with their counterparts from the adjacent subunits, but do not make tight contacts. The ring is stabilized by the RNA belt, and by the “N chain” resulting from insertion of the N-arm of one subunit into the compact fold of the adjacent one. This lateral connectivity helps explain the observed malleability of the N-N interactions in the flexible, yet very stable, RSV nucleocapsid.
Together with electron microscopy data, the structure provides a model for the helical RSV genomic nucleocapsid complex – the viral template for RNA synthesis – with the bases accessible for readout by the polymerase without needing to disassemble the nucleocapsid helix. The model reveals important interaction sites that can be targeted for anti-RSV treatment.
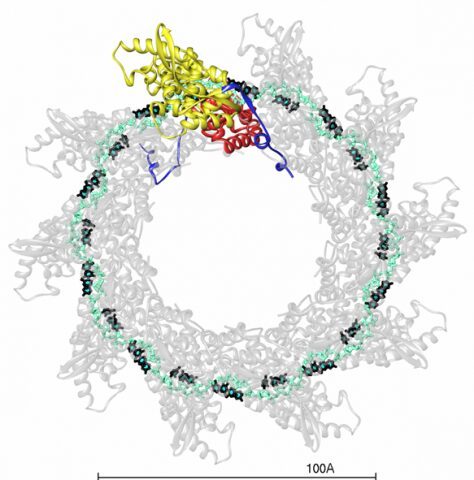
The decameric ribonucleoprotein complex of RSV.
The RNA backbone and bases are displayed in cyan and black respectively. One N-subunit is colored according to domains, with NTD, CTD and arms in yellow, red and blue respectively.
Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagné N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eléouët JF, Rey FA. Science. 2009 Nov 27;326(5957):1279-83.
This work was supported by the sixth European Union Framework Programme consortium, the French ‘Agence Nationale pour la Recherche’ (ANR, program MIME), by Merck-Serono and by recurrent fundings from Institut Pasteur and CNRS. R.G.T. benefits from a Marie-Curie RTN fellowship (EIHCV) and P.F.V. benefited from a postdoctoral fellowship by the French Fondation pour la Recherche Médicale.


