Présentation
The one-dimensional sequence of the genome is carried by long polymers (chromosomes), which fold in the three dimensional volume of the nucleus. How this folding occurs and what it implies for genome function remains largely unknown. To better describe genome architecture in yeast, we developed a method that maps the nuclear territories of fluorescently tagged chromatin loci by computationally analyzing images of thousands of cells [1]. This revealed a strong territorial organization of the yeast nucleus (Figure 1, left). Application of this method to most yeast telomeres allowed us to identify some of the main determinants of locus positioning: genomic distance to centromere and hindrance by the nucleolar compartment [2].
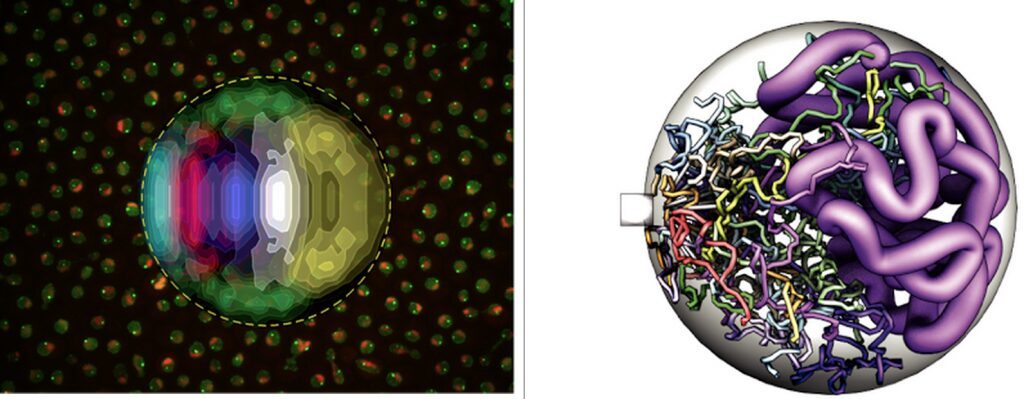
To better understand the mechanisms of chromosome folding, we developed a simple predictive model of chromosome dynamics based on polymer physics (Figure 1, right). The model’s predictions are in very good quantitative agreement with our imaging data and with genome-wide DNA contact frequencies measured biochemically [3]. This suggests that large-scale genome architecture in yeast is governed mainly by generic properties of randomly moving chromosomes rather than by DNA-specific factors. We now explore the consequences of this model for a quantitative understanding of functional processes including DNA repair [4][5] and gene expression. We also investigate the universality of our model’s assumptions by extending it to other organisms. Recently, we started working towards quantitative models of gene expression and RNA processing, and started developing methods to localize and count individual transcripts in single cells [6].
References:
[1] A. B. Berger, G. G. Cabal, E. Fabre, T. Duong, H. Buc, U. Nehrbass, J. C. Olivo-Marin, O. Gadal, and C. Zimmer, “High-resolution statistical mapping reveals gene territories in live yeast,” Nature Methods, vol. 5, no. 12, pp. 1031–1037, 2008.
[2] P. Thérizols, T. Duong, B. Dujon, C. Zimmer, and E. Fabre, “Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres,” Proceedings of the National Academy of Sciences, vol. 107, no. 5, p. 2025, 2010.
[3] H. Wong, H. Marie-Nelly, S. Herbert, P. Carrivain, H. Blanc, R. Koszul, E. Fabre, and C. Zimmer, “A Predictive Computational Model of the Dynamic 3D Interphase Yeast Nucleus.,” Current Biology : CB, vol. 22, no. 20, pp. 1881–90, Oct. 2012.
[4] N. Agmon, B. Liefshitz, C. Zimmer, E. Fabre, and M. Kupiec, “Effect of nuclear architecture on the efficiency of double-strand break repair,” Nature Cell Biology, vol.15, no. 6, pp. 694-9 (2013).
[5] Wong H, Arbona J-M, Zimmer C. “How to build a yeast nucleus.” Nucleus. Aug 22; 4(5):1-6 (2013).
[6] F. Mueller, A. Senecal, K. Tantale, H. Marie-Nelly, N. Ly, O. Collin, E. Basyuk, E. Bertrand, X. Darzacq, and C. Zimmer, “FISH-QUANT: automatically counting transcripts in 3D FISH images,” Nature Methods, Apr; 10(4):277-8, 2013.




