Présentation
Gilles Fischer, Celia Payen
Despite the numerous pathways that aim at preserving genome integrity (DNA recombination and repair, cell cycle checkpoints, etc…), chromosomes are very dynamic structures that accumulate rearrangements over time. These rearrangements (duplications/deletions, translocations, inversions) lead to the modification of both the gene order and the gene content of the genomes and therefore have profound effect onto the evolution of species. We are interested in understanding the phenomenology, the molecular mechanisms as well as the selective impact of chromosomal rearrangements onto genome evolution. We developed a complementary approach between experimental genetics in Saccharomyces cerevisiae and in silico analyses of genomic data from different Hemiascomycetous yeast species.
Our main questions are:
- How gene order is reorganized between genomes of related species?
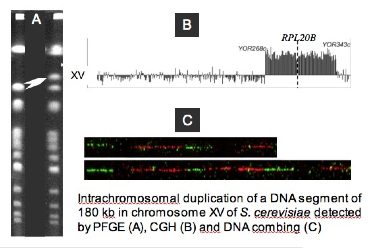
- How segmental duplications are generated in a genome?
 |
 |
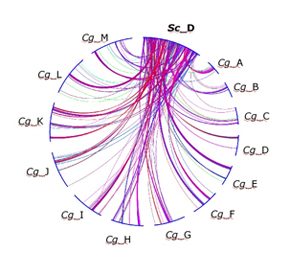
| Schematic representaion of gene order re-assortment between chromosome D of S. cerevisiae (Sc_D) and the 13 chromosomes of C. glabrata (Cg_A to Cg_M) |
