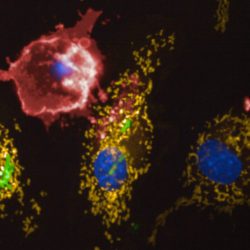
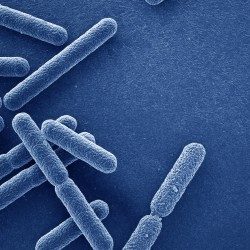
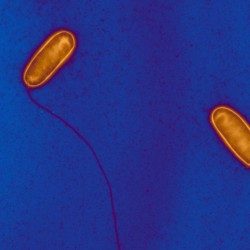
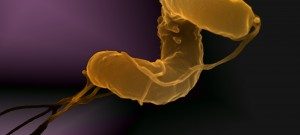
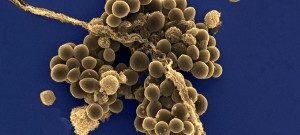
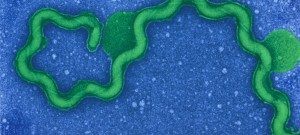
Besides their major role in many infectious diseases, bacteria also serve as models to understand fundamental biological mechanisms. The research performed in the Department of Microbiology mainly focuses on the molecular characterization of functions that enable bacteria to interact with their environment and, in some cases, to cause diseases.
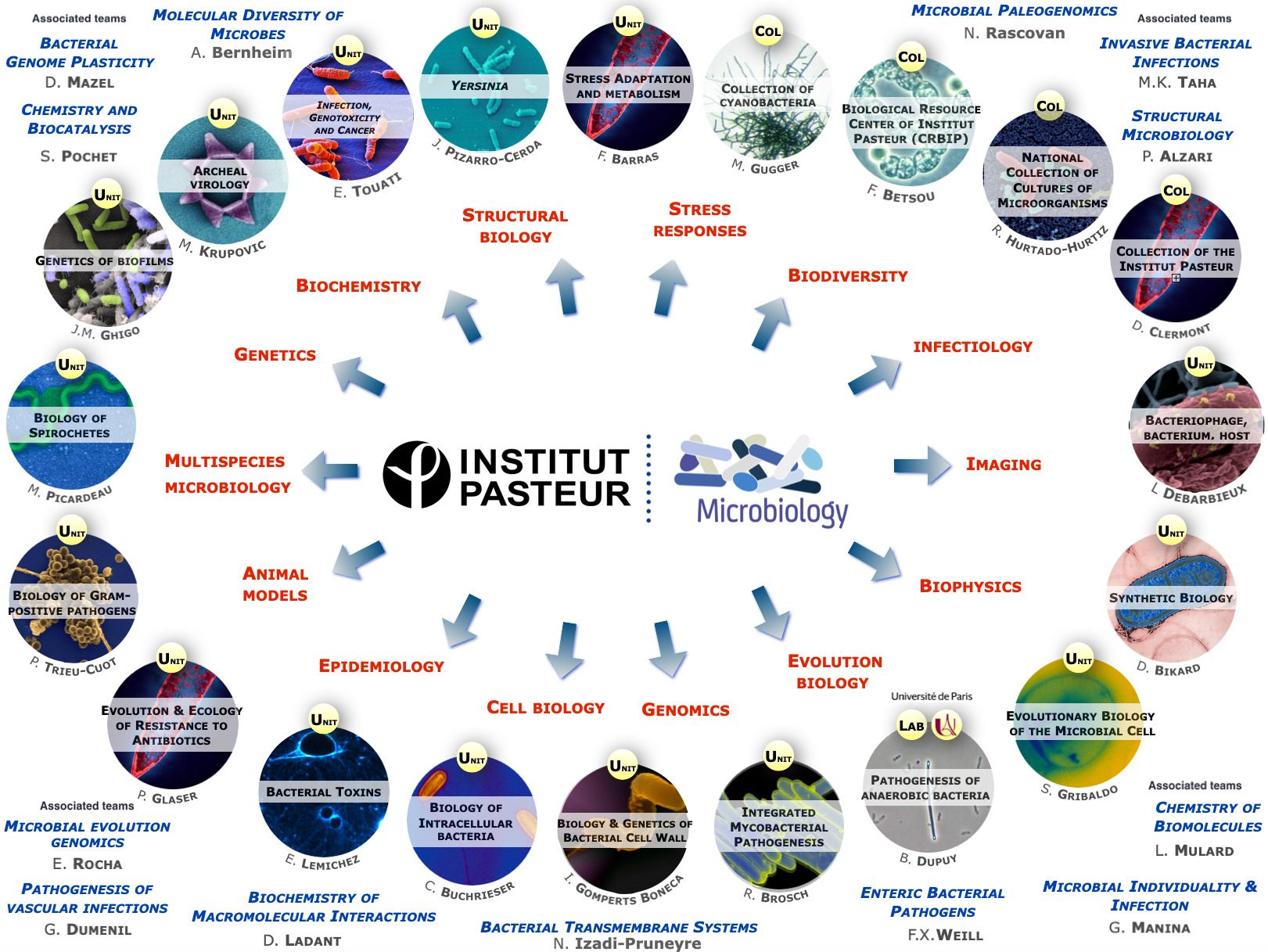
The scientists of the Department of Microbiology study various bacteria and Archaea (and their viruses) as model systems to analyze fundamental biological processes at the population, cellular and molecular levels. They also focus on mechanisms rendering some of these microorganisms virulent and enabling them to evade the host immune system, or to develop resistance to antibiotics. For these studies, the scientists of the Department of Microbiology possess a wide range of expertise and use diverse integrative approaches to improve our understanding of the biology of these microorganisms. These studies also constitute a prerequisite for the development of new therapies or new diagnostic tools that can be used to treat or prevent bacterial infections. The Department of Microbiology includes 20 teams: 15 research teams, 1 Institut Pasteur/Paris University laboratory, and 4 collections. Three entities also host a National Reference Center and 2 are WHO Collaborating Centers. In addition, several research teams from other departments (Genomes & Genetics, Structural Biology and Chemistry and Infection & Epidemiology) are affiliated to the Department of Microbiology. The Department also hosts a joint research entity with the CNRS (French National Center for Scientific Research) – UMR2001.
_______________________
Click to access to the
______________________

(2023) Chromosome folding and prophage activation reveal specific genomic architecture for intestinal bacteria., Microbiome May; 11(1): 111.
Bacterial capsular polysaccharides with antibiofilm activity share common biophysical and electrokinetic properties., Nat Commun May; 14(1): 2553.
Elucidating dynamic anaerobe metabolism with HRMAS 13C NMR and genome-scale modeling., Nat Chem Biol May; 19(5): 556-564.
Legionella para-effectors target chromatin and promote bacterial replication., Nat Commun Apr; 14(1): 2154.
Cas9 off-target binding to the promoter of bacterial genes leads to silencing and toxicity. , Nucleic Acids Res Mar; (): .
Yersiniomics, a Multi-Omics Interactive Database for Yersinia Species., Microbiol Spectr Feb; (): e0382622.
Microbiota-induced active translocation of peptidoglycan across the intestinal barrier dictates its within-host dissemination., Proc Natl Acad Sci U S A Jan; 120(4): e2209936120.
Bioenergetic State of Escherichia coli Controls Aminoglycoside Susceptibility., mBio Jan; (): e0330222.